Figure 2. Examples of the HTS-HTS data analyses.
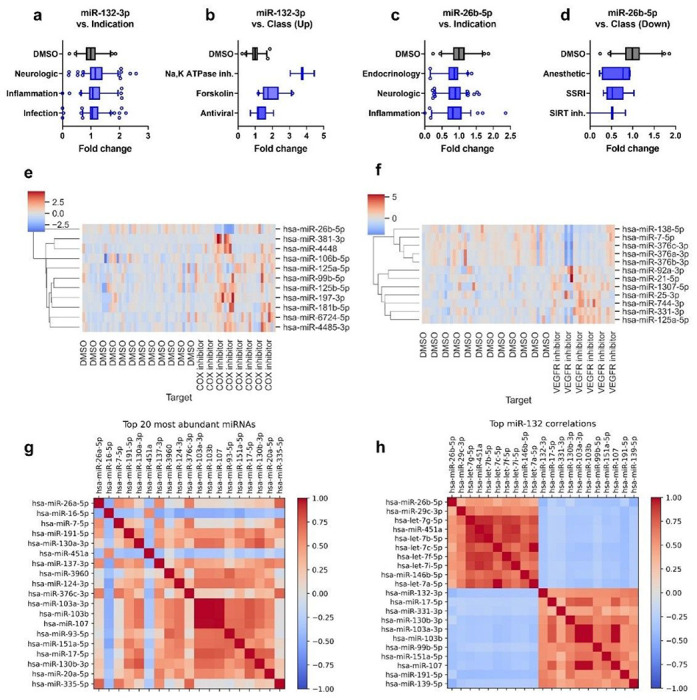
a-d The dataset can be explored to identify classes of compounds regulating a miRNA of interest, for example, miR-132 and miR-26b, which are dysregulated in ADRD. a, miR-132 is mildly upregulated by compounds in clinical use for treating neurological diseases, inflammation, and infection. b, miR-132 is upregulated by compounds classified as Na, K-ATPase inhibitors, forskolin (positive control), and antiviral. c, miR-26b is mildly downregulated by compounds in clinical use for treating endocrinologic and neurological diseases and inflammation d, miR-26b is downregulated by compounds classified as anesthetics, selective serotonin reuptake inhibitors (SSRI), and SIRT inhibitors. e-f, The dataset can be used to explore what miRNAs may be regulated by specific classes of compounds, for example, through clustered heatmaps of miRNAs altered by COX inhibitors (N=33) or VEGFR inhibitors (N=23) compared to DMSO controls (N=46). g-h, The dataset can also be used to explore relationships between miRNAs in human iPSC-derived neurons through correlation maps among a group of miRNAs, such as (g)the top abundant miRNAs or (h) miRNAs with top negative and positive correlation to miR-132.
