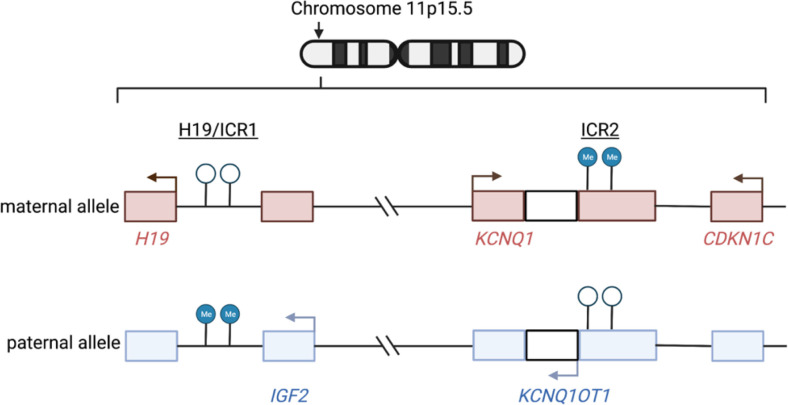
Figure 1. Mechanisms of dysregulated imprinting at chromosome 11p15.5.
Chromosome 11p15.5 houses a cluster of imprinted genes (including IGF2, KCNQ1, CDKN1C) and noncoding RNAs (including H19, KCNQ1OT1) that are either expressed only from the maternal or paternal allele. The expression of these genes is regulated by differential methylation at two imprinting control regions (ICR) – 11p15.5 H19/ICR1 and KCNQ1OT1/ICR2. In normal physiology, H19/ICR1 is methylated on the paternal allele only and IGF2is expressed from this allele. In contrast, ICR2, located at the promoter region of KCNQ1OT1 is methylated on the maternal allele only, leading to expression of KCNQ1 and CDKN1C from the maternal allele. The normal physiologic imprinting status at 11p15.5 is referred to as 11p15.5 retention of imprinting (ROI) throughout this manuscript. Two predominant mechanisms occur that disrupt imprinting and result in biallelic expression of IGF2: 1) 11p15.5 H19/ICR1 loss of imprinting (LOI) refers to site-specific epigenetic gain of methylation at H19/ICR1 and 2) 11p15.5 copy neutral loss of heterozygosity (cn-LOH) refers to genetic deletion of the maternal allele and duplication of the paternal allele (paternal uniparental disomy). With 11p15.5 LOI, there is hypermethylation of H19/ICR1 and normal methylation of KCNQ1OT1/ICR2. With 11p15.5 LOH, there is hypermethylation of H19/ICR1 and hypomethylation of KCNQ1OT1/ICR2. Arrows indicate active transcription reflective of the normal physiologic imprinting pattern. Graphic made with biorender.com