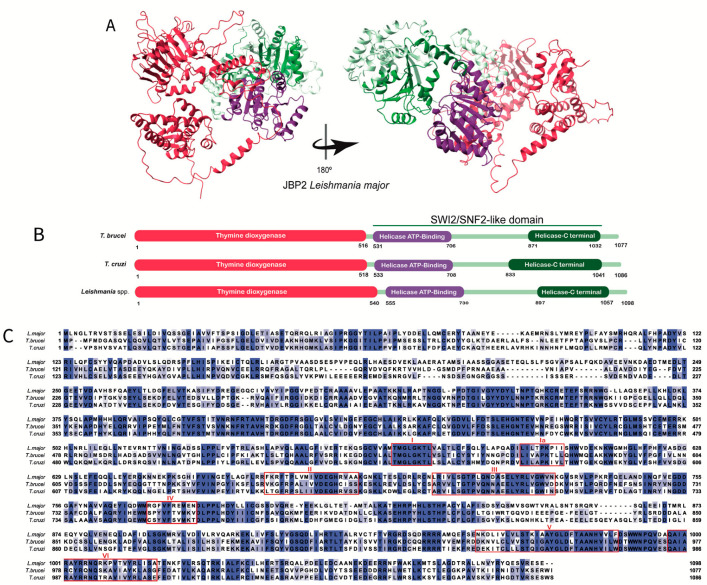
Figure 3.
The JBP2′s structure: (A) Tridimensional Leishmania major’s JBP2 predicted structure obtained in the AlphaFold database (Q4QFY1 (JBP2_LEIMA). The conserved thymine dioxygenase domain (red) is depicted. The SWI/SNF2 domain is highlighted, and the subdomains (the helicase ATP-binding termianl and helicase C-terminal) are shown in purple and green, respectively. (B) The domain’s positions in the amino acid sequence obtained in the UniProt database for Trypanosoma brucei (Q57X81), Trypanosoma cruzi (Q4DCH3), and Leishmania major (Q4QFY1) are highlighted. The conserved thymine dioxygenase domain is shown in red. The SWI/SNF2 is depicted, and the subdomains (the helicase ATP- binding and helicase C-terminal) are shown in purple and green, respectively. (C) Multiple alignments were performed using the amino acid sequences obtained in the UniProt database for Trypanosoma brucei (Q57X81), Trypanosoma cruzi (Q4DCH3), and Leishmania major (Q4QFY1). The amino acids highlighted in different tones of blue, from light to dark blue, represent the percentage of identity (20–100%). The two histidines and the aspartic acid residues involved in Fe2+ binding and the arginine residue important for binding 2-oxoglutarate are restricted by a black box. The conserved motifs (I, Ia, II, III, IV, V, and VI) in the SWI/SNF2 domain are restricted by individual red boxes [34].