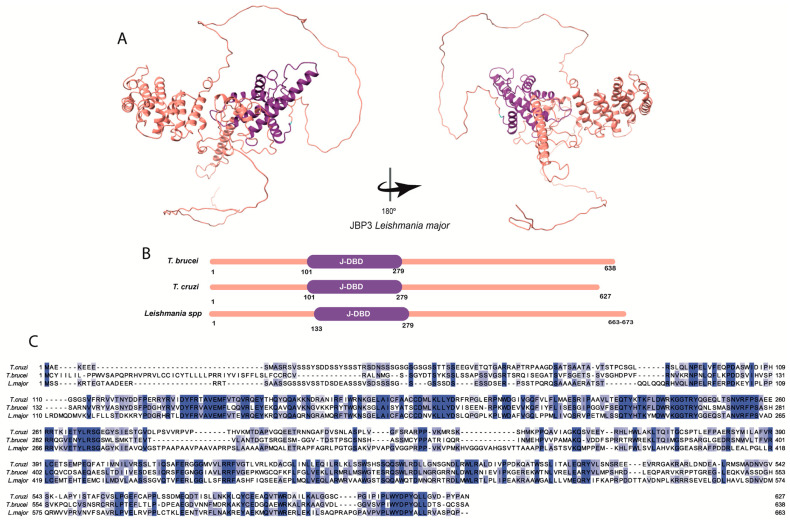
Figure 4.
The JBP3′s structure: (A) tridimensional Leishmania major’s JBP3 predicted structure obtained in the AlphaFold database (Q4Q239). The conserved J-DBD domain (purple) is depicted. (B) The domain’s positions in the amino acid sequence obtained in the UniProt database for L. major (Q4Q239), Trypanosoma brucei (Q38BC1), and Trypanosoma cruzi (Q4CUX1) are highlighted. The conserved J-DBD domain is shown in purple. (C) Multiple alignments were performed using the amino acid sequences obtained in the UniProt database for L. major (Q4Q239), Trypanosoma brucei (Q38BC1), and Trypanosoma cruzi (Q4CUX1). The amino acids highlighted in different tones of blue, from light to dark blue, represent the percentage of identity (20–100%).Furthermore, the degree of conservation (identity versus similarity) between the three JBPs from the aligned species is low (about 32% for JBP1 and JBP2 and 19% for JBP3) (Table 1 and Figure 2C and Figure 3C). However, they all preserve the structural and functional domains involved in J synthesis and J-binding to DNA, indicating that the proteins retain their physicochemical properties, potentially allowing them to function similarly.