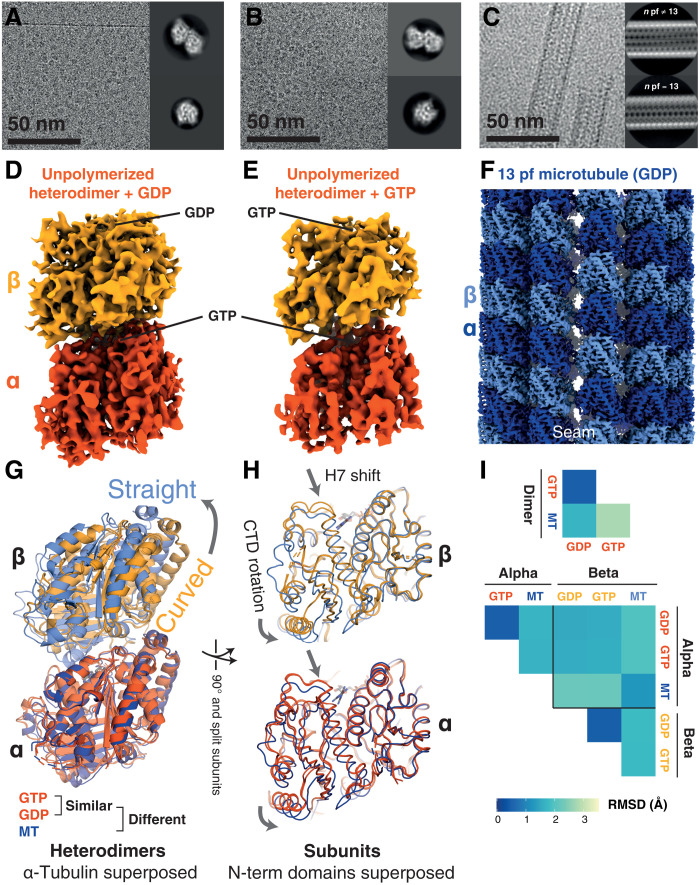
Fig. 3. Cryo-EM structures of free/un-complexed D. melanogaster (Dm) tubulin heterodimers and an MT recapitulate classical tubulin assembly switches and reiterate the absence of a nucleotide state-driven switch.
(A) Cryo-EM study of Dm tubulin heterodimers prepared with GDP. Representative micrograph and 2D classes. Processing scheme can be found in fig. S3. (B) Cryo-EM study of Dm tubulin heterodimers prepared with GTP. Representative micrograph and 2D classes. Processing scheme can be found in fig. S4. (C) Cryo-EM study of Dm MTs prepared with GTP. Representative micrograph and 2D classes. (D and E) Cryo-EM maps of Dm tubulin prepared with GDP or GTP occupying the β-tubulin binding site as indicated. (F) Cryo-EM map of Dm tubulin polymerized into 13 protofilament MT. (G and H) Comparison of Dm models (PDB; GTP: 7QUD, GDP: 7QUC, and MT: 7QUP) from the dimer maps (orange: dark—α subunit, light—β subunit) and the 13-protofilament MT (blue shades). Structures in (G) are aligned on the N-terminal domain of α-tubulin; structures in (H) are aligned on the N-terminal domains of the respective subunits. (I) Cα RMSD comparison of dimers (top) and individual subunit structures (bottom), following superposition as in (G) and (H). Unpolymerized heterodimers are very similar to one another.