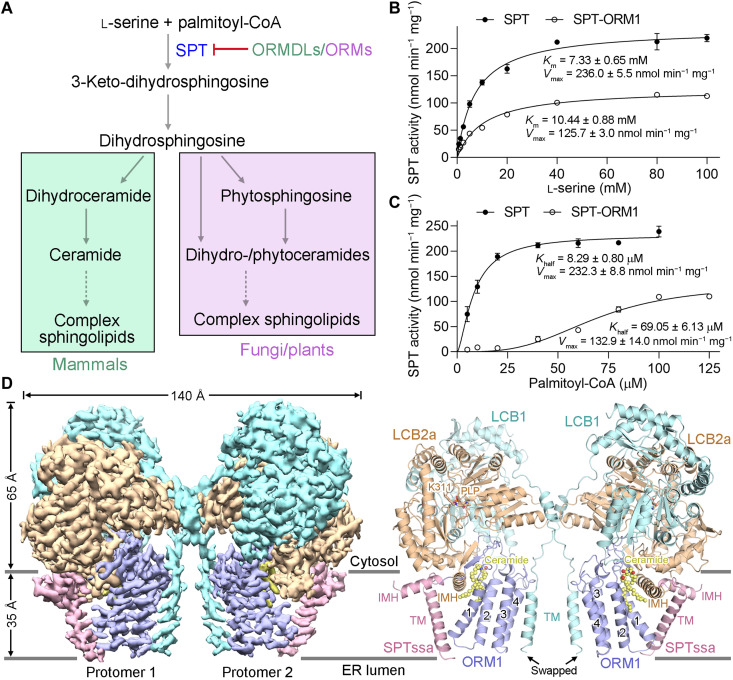
Fig. 1. Biochemical characterization and structural determination of AtSPT-ORM1 complex.
(A) A diagram of the de novo sphingolipid biosynthetic pathways in mammals, fungi, and plants. The serine palmitoyltransferase (SPT) activity is negatively regulated by ORMDLs/ORMs. (B) SPT activity versus l-serine concentration measured using SPT or SPT-ORM1 complex. The data points were fitted with a Michaelis-Menten equation. For all curves and dot plot graphs, each data point is the average of three independent experiments, and error bars indicate the SEM. (C) SPT activity versus palmitoyl–coenzyme A (CoA) concentration for SPT and SPT-ORM1 complex. The data points were fitted with an allosteric sigmoidal equation. (D) The cryo–electron microscopy (cryo-EM) map and overall structure of SPT-ORM1 complex. LCB1 and LCB2a are shown in light cyan and wheat, respectively; SPTssa and ORM1 are shown in pink and light blue, respectively; ceramide is shown in yellow. The cofactor pyridoxal 5′-phosphate (PLP) is shown in wheat sticks. TM, transmembrane helix; IMH, in-plane membrane helix; ER, endoplasmic reticulum. All structural figures were prepared using PyMOL (59) or UCSF Chimera (60).