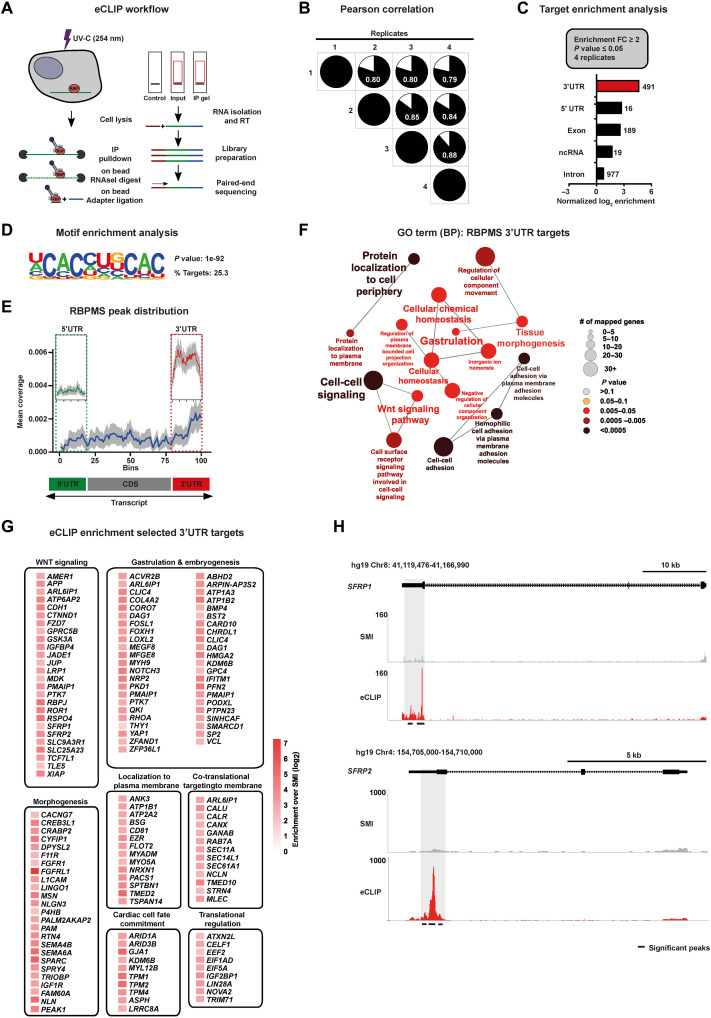
Fig. 4. mRNAs encoding cardiac mesoderm regulators are targeted by RBPMS via 3′UTR binding.
(A) Schematic of the eCLIP-seq approach used to faithfully generate a transcriptome-wide direct binding map for RBPMS at single-nucleotide resolution. (B) Biological quadruplicates of RBPMS eCLIP-seq show at least 80% overlap. Pie charts show the correlation of statistically significant uniquely mapped reads for each replicate over SMInput. (C) RBPMS reliably binds predominantly the 3′UTR of transcripts, demonstrated here by the distribution of the significantly enriched eCLIP peaks against the paired SMInput (FC ≥ 2; P ≤ 0.05 in all four replicates). (D) Top sequence motif significantly bound by RBPMS. (E) Metagene plot visualizing the RBPMS peak distribution over SMInput illustrating prominent 3′UTR binding. (F) 3′UTR targets of RBPMS regulate molecular processes central to mesoderm/cardiac commitment, including WNT signal transduction, depicted by significantly enriched GO terms. (G) A curated set of RBPMS 3′UTR targets grouped based on their proven role in the indicated cellular, developmental, and functional process, depicted as a heatmap of fold enrichment over SMInput. (H) Representative read density tracks show read density for RBPMS across the gene body of SFRP1 and SFRP2, a representative target.