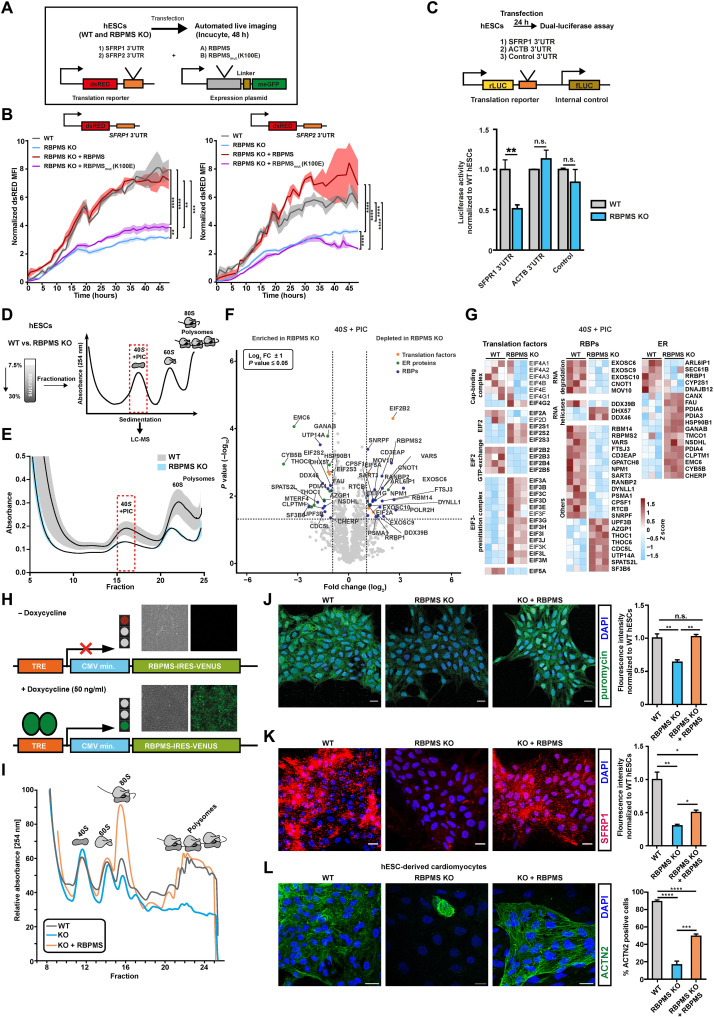
Fig. 6. RBPMS specializes mRNA translation in pluripotency, selectively via 3′UTR binding and globally by controlling translation initiation and ribosome recruitment.
(A) Schematic of the reporter system and the experimental workflow used to investigate the 3′UTR binding motif–dependent regulation of translation by RBPMS in hESCs. (B) RBPMS activates translation of reporter mRNA carrying RBPMS binding motifs in the 3′UTR, evaluated by time-lapse microscopy. (C) The presence of the RBPMS binding motif is required for 3′ binding–dependent translation activation by RBPMS evaluated using indicated luciferase-based bicistronic reporters. (D) Schematic outlining the translation complex profiling–based isolation of 40S and preinitiation complex (PIC), followed by proteomics analysis in WT and RBPMS-KO hESCs. (E) Translation complex profiling traces of WT and RBPMS-KO hESCs (shades represent SEM). 40S + PIC fractions were subjected to LC-MS/MS. (F) Proteins significantly changing in the 40S + PIC fraction between WT and RBPMS-KO hESCs represented as volcano plot. Dashed lines indicate significance thresholds (−log10 P ≥ 1.3 and log2 FC ± 2) (selected translation factors, ER proteins, and RBPs are highlighted by orange, green, and blue dots, respectively). (G) Heatmap depicting differentially enriched translation factors, RBPs, and translation-associated ER proteins in the 40S + PIC fraction between WT and RBPMS-KO hESCs (significantly changing proteins are highlighted in bold). (H) Illustration of the PiggyBac-based strategy used to reexpress RBPMS in RBPMS-KO. Representative microscopy images in the inlets before and after induction. Timely reconstitution of RBPMS in RBPMS-KO hESCs rescues (I) translation defects, (J) protein synthesis defects, (K) translation defect of representative 3′UTR target of RBPMS, SFRP1, and (L) cardiac differentiation defect. Quantification of the microscopy images on the right side as bar graphs. Error bars represent ±SEM; P values calculated using Student’s t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n = 3).