Summary
How canonical cytokinesis is altered during germ cell division to produce stable intercellular bridges called ring canals is poorly understood. Here, using time-lapse imaging in Drosophila, we observe that ring canal formation occurs through extensive remodeling of the germ cell midbody, a structure classically associated with its function in recruiting abscission-regulating proteins in complete cytokinesis. Germ cell midbody cores reorganize and join the midbody ring rather than being discarded, and this transition is accompanied by changes in centralspindlin dynamics. The midbody-to-ring canal transformation is conserved in the Drosophila male and female germlines and during mouse and Hydra spermatogenesis. In Drosophila, ring canal formation depends on Citron kinase function to stabilize the midbody, similar to its role during somatic cell cytokinesis. Our results provide important insights into broader functions of incomplete cytokinesis events across biological systems, such as those observed during development and disease states.
Graphical Abstract
eTOC blurb
Ring canals connect germ cells in syncytial groups and are necessary for gamete development. Price et al show that ring canals form through remodeling of germ cell midbodies in Drosophila, Hydra, and mouse. Perturbation of midbody formation and subsequent reorganization into ring canals affects communication between germ cells.
Introduction
In contrast to canonical cell division, animal germline cells and certain somatic cells undergo incomplete cytokinesis to produce cells in syncytial groups that remain connected by intercellular bridges, often called ring canals. These bridges are highly conserved in male and female germline cells from sponges to mammals1–8. Ring canals are composed of many cytokinesis proteins revealing the intimate relationship between cytokinesis and ring canal formation1. While it is clear that inhibition of membrane abscission is necessary for incomplete cytokinesis, it remains unknown how cytokinesis is altered during germ cell division to give rise to ring canals. In particular, it is unclear how former contractile ring and cytokinesis proteins come to be enriched in the membrane-attached stable ring canal and how these cytoskeletal changes are coordinated with abscission inhibition.
Complete cytokinesis is orchestrated by a specific organelle, the midbody, that resides at the center of the transient intercellular bridge between nascent cells9. Constriction of the contractile ring compacts the central spindle microtubules and their associated proteins to form the midbody, the organization of which can be further subdivided: 1) the midbody core, the central region of the midbody that contains the centralspindlin complex; 2) the midbody ring that surrounds the midbody core and is comprised of contractile ring proteins including Citron kinase and Anillin; and 3) the midbody arms that flank the midbody core and contain Aurora B and components of the chromosome passenger complex (see D’Avino10 for review). The midbody serves to recruit microtubule-severing proteins and the membrane abscission machinery needed to finally separate new sister cells11–13.
Midbodies have also been implicated in incomplete cell division resulting in ring canals. By electron microscopy, ring canals are characterized by an electron-dense limiting membrane that resembles the cleavage furrow at the midbody stage of cytokinesis14,15. In the mouse male germline, loss of the ring canal-promoting factor TEX14, which blocks abscission through competitive inhibition of the ESCRT-associated protein CEP-5516, results in the formation of midbody-like foci between dividing cells, implying that midbody formation is a normal step in ring canal biogenesis17. However, a TEX14-based mechanism to block abscission is restricted to mammals since more primitive animal genomes do not encode CEP-55. A different model has emerged from studies of fixed Drosophila cells in which ring canals form due to arrested constriction of the contractile ring before midbody formation as midbodies have not been observed in dividing germ cells1,6,8,18–20.
The temporal aspects of ring canal formation have not been described and it remains unclear how midbody or contractile ring behavior during incomplete cytokinesis contributes to the formation of a stable ring canal. Furthermore, compositional differences between Drosophila male and female germline ring canals and between mouse and Drosophila male ring canals, combined with differences in proposed models of ring canal formation, raise the question of whether there is one conserved mechanism of ring canal formation.
To gain new insight into the mechanism of ring canal formation, we performed extensive live imaging to monitor the dynamics of ring canal proteins during incomplete mitosis and subsequent ring canal formation. We imaged several cleavage furrow and ring canal proteins, including the highly conserved MKLP1/kinesin-6 subunit of the centralspindlin complex. In both the Drosophila testis and the ovary, we discovered that ring canal formation is preceded by the formation of a robust midbody intermediate that reorganizes to form a ring with an open lumen. By analyzing MKLP1/kinesin-6 homologs in the mouse and Hydra vulgaris testis, we find that a midbody-to-ring canal transition is a conserved feature of ring canal formation across evolution. Further, we demonstrate that ring canals are the product of stabilized midbody rings. Mechanistically, we found that perturbation of Citron kinase caused defects in midbody organization/dynamics resulting in a delayed or failed midbody-to-ring canal transition and slowed intercellular communication. Our results identify a previously unknown, but highly conserved midbody behavior during programmed incomplete cytokinesis and new insights into the mechanisms that promote ring canal formation.
Results
Drosophila ring canal formation during gametogenesis occurs via a midbody-like intermediate
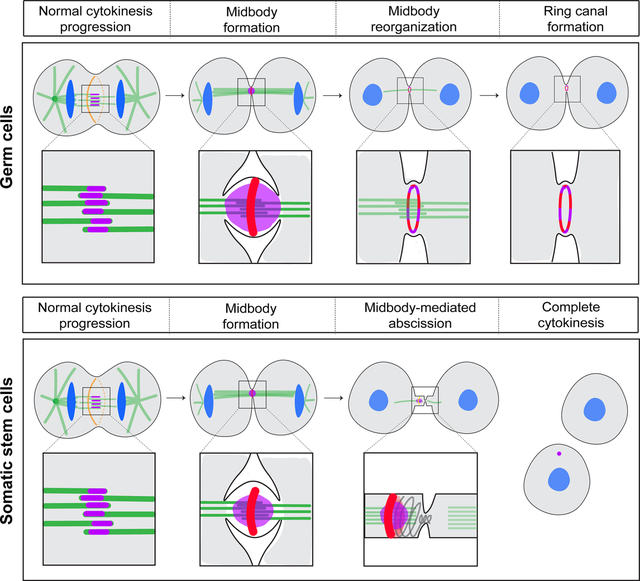
To investigate how germline ring canals are formed, we performed time-lapse confocal imaging of known ring canal components during incomplete mitosis in the Drosophila male germline (Figure 1A). We focused on the highly conserved MKLP1/kinesin-6 protein (known as Pavarotti or Pav in Drosophila and MKLP1/KIF23 in mammals), a subunit of the centralspindlin complex and component of all characterized ring canals1,6. In Drosophila, Pav has also been shown to localize to the central spindle and nascent cleavage furrow21. We imaged either endogenously-tagged Pav::mCherry (see Materials and Methods) or a Pav::GFP transgenic protein expressed at endogenous levels22. To correlate ring canal formation with cell cycle timing, we visualized Pav::mCherry in combination with GFP::Histone. Live imaging revealed that, during anaphase, Pav::mCherry localized to the cleavage furrow and central spindle as expected, consistent with its role in cytokinetic furrowing (Figure 1B, t=0 min, CF). Rather than a cleavage furrow arrest before midbody formation as predicted by one model for ring canal formation in Drosophila (reviewed in Haglund et al.1), we observed formation of a dense midbody core focus containing Pav::mCherry that suggests full constriction of the contractile ring (Figure 1B, t=12 min). Remarkably, rather than being lost as happens during complete cytokinesis, the midbody core focus persisted for approximately 25 minutes and then resolved into a ring with an open lumen (Figure 1B, t=34 minutes). The central spindle microtubules remained present during the midbody-to-ring transition (Figure S1) providing evidence that microtubule severing did not occur during this time.
Figure 1. Drosophila ring canal formation during gametogenesis occurs via a midbody-like intermediate.
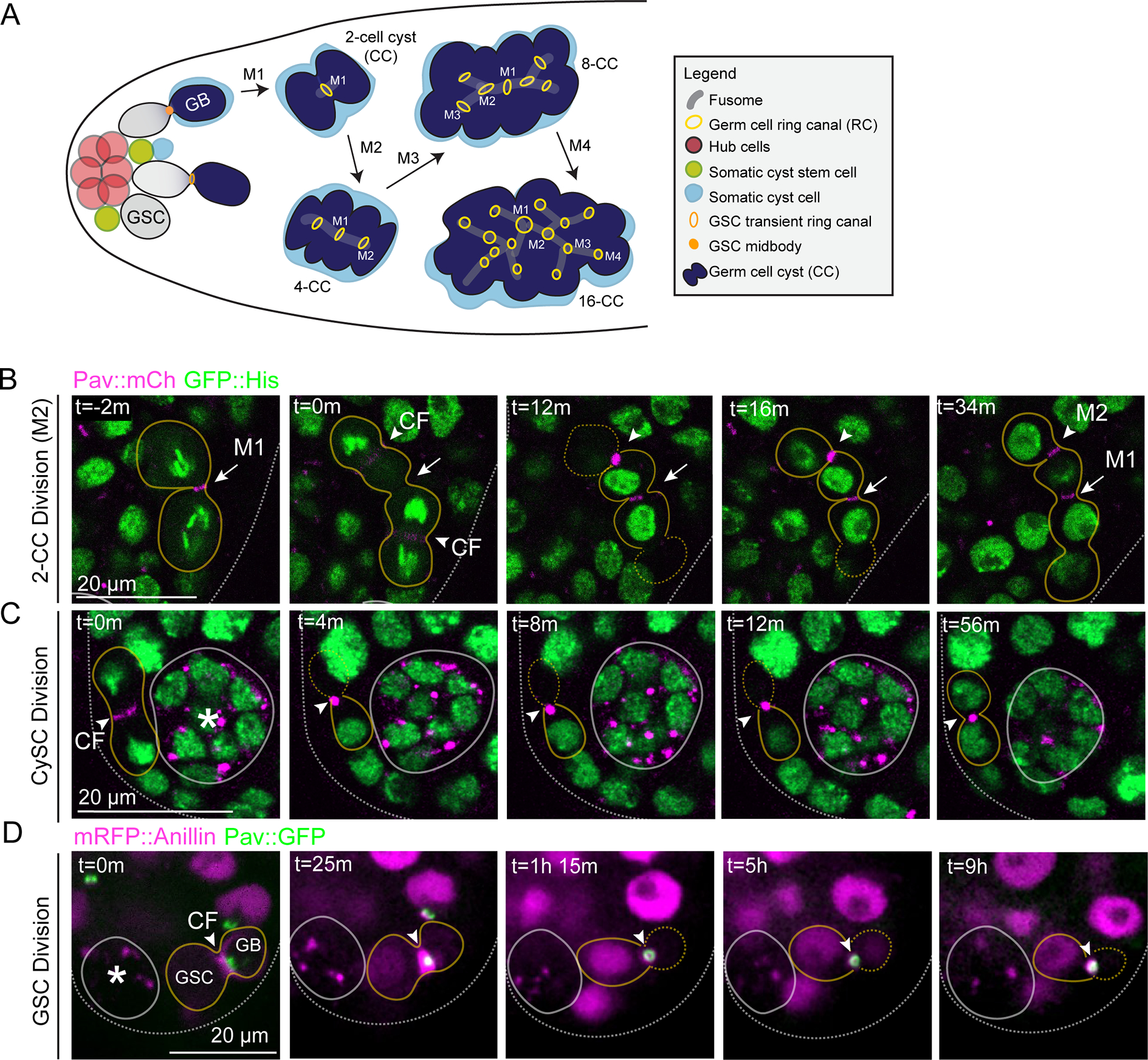
(A) Diagram of Drosophila testis. (B-C) Time-lapse imaging of Pav::mCherry and GFP::Histone. Images were acquired every 2 minutes. See video S1. (B) Still from a representative germ cell cyst division (outlined, yellow; n=49). Arrow represents the ring canal from the first mitotic division (M1); arrowhead represents the cleavage furrow (CF) and resulting ring canal of the second mitotic division (M2). (C) Cyst stem cell division (outlined, yellow; n=8). Arrowhead represents the cleavage furrow and midbody; asterisk, stem cell niche. In (B) and (C), each image is a 2–4 μm Z plane projection. (D) Live imaging of Pav::GFP and mRFP::Anillin during the germline stem cell division (n=5). Asterisk marks the stem cell niche. GSC, germline stem cell; GB, gonialblast. Each image is a single Z plane. Images were acquired every 25 minutes. (B-D) m, min; h, hour.
The presence of Pav in a midbody core and later in a ring is similar to the described localizations of MKLP1 in the midbody core and midbody ring in fixed human cells23,24 suggesting that our observations of Pav reorganization in male germline cells may not be specific to cells undergoing incomplete cytokinesis. To gain insight into the nature of Pav during complete cytokinesis, we analyzed cells that undergo abscission, namely the somatic cyst stem cells (CySC) and germline stem cells (GSC) (Figure 1C, 1D). The CySCs divide asymmetrically to self-renew and produce daughter cyst cells that envelop the gonialblast25–27. We identified dividing CySCs by the orientation of their cleavage planes (parallel to the hub versus perpendicular). We observed two striking differences compared to germ cell divisions: smaller Pav-containing midbodies and no apparent reorganization into Pav-containing rings (Figure 1C, Figure S2A–E). These data suggest that CySC midbodies do not undergo a midbody-to-ring canal transition and the midbodies function canonically to orchestrate membrane abscission.
During male GSC division, cytokinesis is delayed during which many GSCs remain attached to their gonialblast (GB) daughter cells by transient ring canals25 (Figure 1A). The GSC-GB attachment lasts through the next S phase and abscission does not occur until G2 of the following cell cycle28. Furthermore, previous work from Lenhart and DiNardo34 revealed that myosin-II-containing midbody rings persist for ~8.5 hours before condensing to a midbody focus, only after which time abscission occurred. We performed long-term imaging of Pav::GFP and transgenic mRFP::Anillin or Sqh::GFP (Figure S2) to monitor dividing GSCs. We observed that, similar to our observations during incomplete cytokinesis of germ cells, Pav::GFP formed a bright focus following constriction of the contractile ring (Figure 1D). mRFP::Anillin localized to the ingressing cleavage furrow and then to a midbody ring around the Pav::GFP-labeled midbody (Figure 1D, t=25 minutes, respectively). Pav::GFP then reorganized into a transient ring that colocalized with the midbody ring (Figure 1D, t=1 hr and 15 min, Figure S2F–J). Approximately 9 hours after initial cleavage furrow ingression, Pav::GFP and mRFP::Anillin or Pav::GFP and Sqh::GFP once again co-localized in a smaller midbody structure (Figure 1D, t=9 hr, Figure S2K–M), similar to the timing of mature midbody formation observed by Lenhart and DiNardo34. These data suggest germline cell division includes formation of a ring canal that is transient in germline stem cells and stabilized in developing germ cells.
The centralspindlin complex undergoes changes in abundance and organization during ring canal formation
We quantified the fluorescence intensity of Pav-labeled midbodies, midbody diameters and, when applicable, the size of the resultant ring canals in dividing germ cells, cyst stem cells and germline stem cells (Figure 2A–C). From midbody to ring formation in dividing germ cells, the fluorescence intensity of Pav diminished, suggesting a period of midbody maturation (Figure 2A, 2B). Pav mean pixel intensity during incomplete cytokinesis was highest in the nascent midbody focus and underwent a ~1.2-fold reduction in fluorescence intensity over a period of 10 minutes (Figure 2A, t=6 minutes to t=17 minutes; Figure 2B, 10 to 20 min.). Resolution to a nascent ring canal with an open lumen was accompanied by an additional 2-fold reduction in fluorescence (Figure 2A, t=17 minutes to t=30 minutes; Figure 2B, 20 to 30 minutes). In contrast to the marked reduction in Pav protein during midbody remodeling in germ cells, Pav protein levels in the hub cell foci remained mostly static (Figure 2B), revealing that the decrease in Pav levels during ring canal formation was not due to photobleaching. Furthermore, imaging of photoconverted Pav::Dendra2 (driven by nanos-Gal4) in dividing germ cells recapitulated the changes in Pav fluorescence intensity during ring canal formation and revealed that ring canal formation is the direct result of midbody reorganization (Figure S3).
Figure 2. The centralspindlin complex undergoes changes in abundance and organization during ring canal formation.
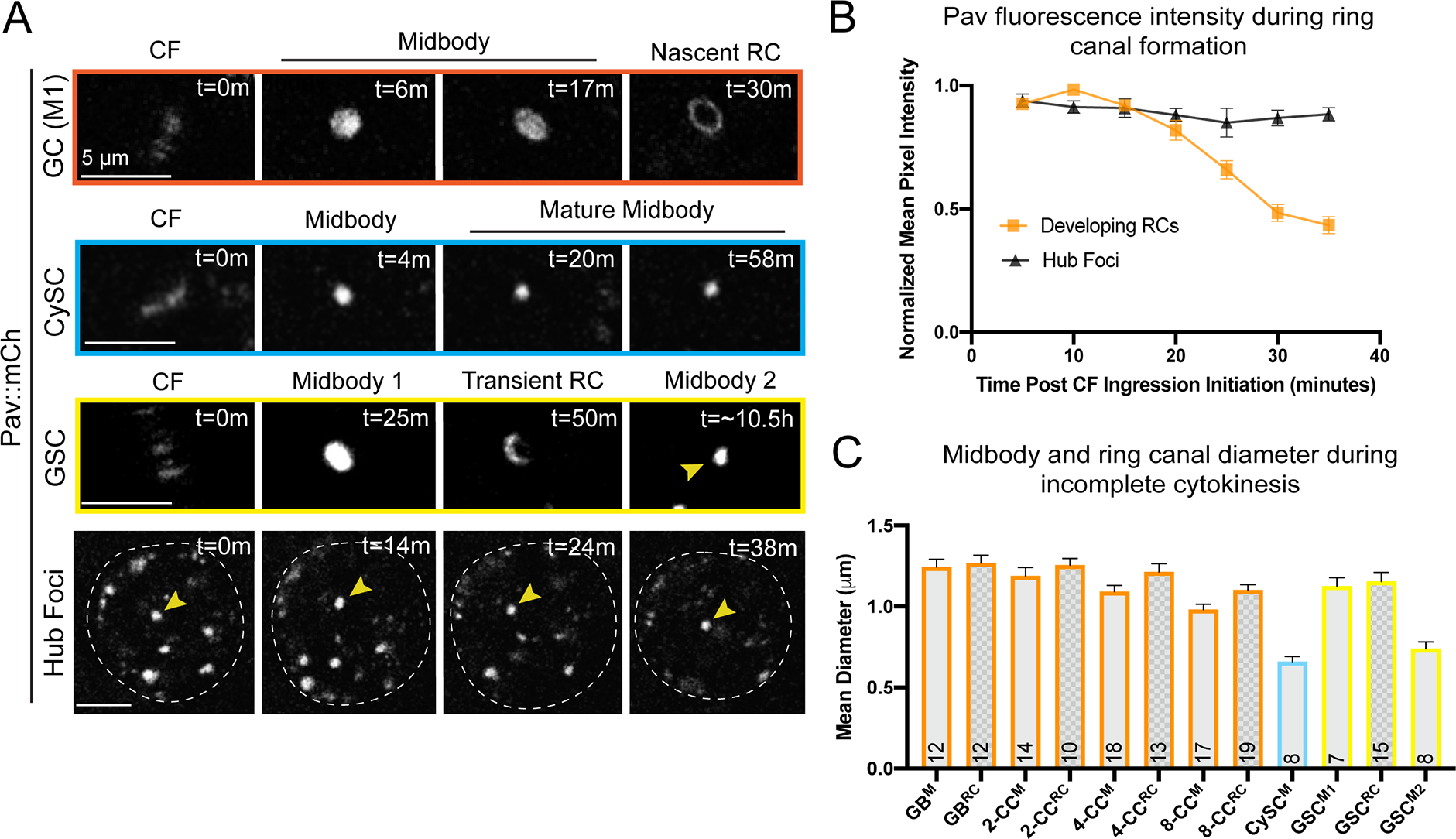
(A) Time-lapse imaging of Pav::mCherry. GC, germ cell; M1, first mitotic division (n=12); CySC, cyst stem cell (n=8); GSC, germline stem cell (n=7); m, minutes. Each panel is a 2–4 μm Z projection. Pav localizes in the midbody core (labeled midbody) before reorganizing to form a ring (nascent RC or transient RC) in germ cells and germline stem cells, but not cyst stem cells. (B) Pav::mCherry normalized mean pixel intensities during ring canal formation in dividing germ cells as in (A). Mean pixel intensities (±SEM) of Pav::mCherry in a 2 μm2 region-of-interest were normalized to the mean pixel intensities of the same ROI in the germ cell midbody (orange) or to the hub foci at t=0m (black) (n=16). (C) Midbody diameters and resulting ring canals were measured in dividing cells of the testis shown in (A). GB, gonialblast; CC, cell cyst; M, midbody; RC, ring canal; M1, the midbody that resolves to form a transient ring canal; M2, the second, smaller midbody that forms prior to germline stem cell abscission. The sample size for each midbody and RC measured is listed in each bar. The average diameter is listed (±SEM). Midbody diameters were determined from measurements of the full width at half maximum and ring canal diameters determined from the distance between the two highest pixel intensities of a line scan. The mature midbodies that form in the CySC (CySCM) and GSC (GSCM2) are not statistically different from each other (p=0.1519, unpaired two-tailed t-test), but are statistically different from the germ cell midbodies (GB, 2-. 4-, 8-CC M) (ordinary ANOVA, p<0.0001). Germ cell midbody diameters are statistically different from each other (ordinary ANOVA, p=0.0190). Germ cell ring canals are also statistically different from each other (ordinary ANOVA, p=0.0190).
The diameters of midbodies differed in cells that did or did not form ring canals. Midbodies that formed rings in dividing germ cells (Figure 2A: GC) or during delayed abscission of GSCs (Figure 2A: GSC, Midbody 1) were about 1.2 μm in diameter (Figure 2C). In contrast, midbodies in CySCs (Figure 2A: CySC) and the mature midbodies in GSCs (Figure 2A: GSC, Midbody 2) had average diameters of 0.6 and 0.7 μm, respectively (Figure 2C), which is still large enough to resolve a transition to a ring if it happened (Figure S2). These data suggest the possibility that midbody size must be large enough to support the remodeling process that results in a ring shape.
Based on our live imaging of Pav::mCherry, we defined four stages of ring canal biogenesis that are accompanied by distinct changes in Pav levels and organization: 1) central spindle formation/cleavage furrow ingression; 2) midbody formation; 3) midbody maturation; and 4) nascent ring canal formation.
A midbody-to-ring reorganization is conserved in dividing Drosophila female germ cells and in distantly related male germ cells
Evidence supporting the model of contractile ring arrest in the formation of ring canals came from studies in the Drosophila female germline. Mutations in the myosin-binding subunit of myosin light chain phosphatase, which negatively regulates Myosin II-mediated contraction, resulted in small ring canals that were attributed to over-constriction of the contractile ring20. This study provided genetic evidence implicating proteins necessary for contractile ring constriction in ring canal formation. However, the phenotype was specific to the female germline raising the question of whether different mechanisms drive ring canal formation during spermatogenesis and oogenesis.
To address this question, we examined Pav::GFP behavior during Drosophila oogenesis using live imaging of germaria where incomplete mitoses occur. We found the same midbody-to-ring reorganization that we observed in male germ cell division in the testis (Figure 3A, Figure S4). The average time from anaphase onset to ring canal formation was 50 minutes (n=3), which was slower than in the testis. Consistent with previous quantifications of ring canal size29, we found that midbody diameter and nascent ring canal size were smaller with each successive division as cysts moved posteriorly in the germarium (Figure S4). These data suggest a common mechanism of ring canal formation during Drosophila gametogenesis in males and females.
Figure 3. A midbody-to-ring reorganization occurs in dividing Drosophila female germ cells and in distantly related male germ cells.
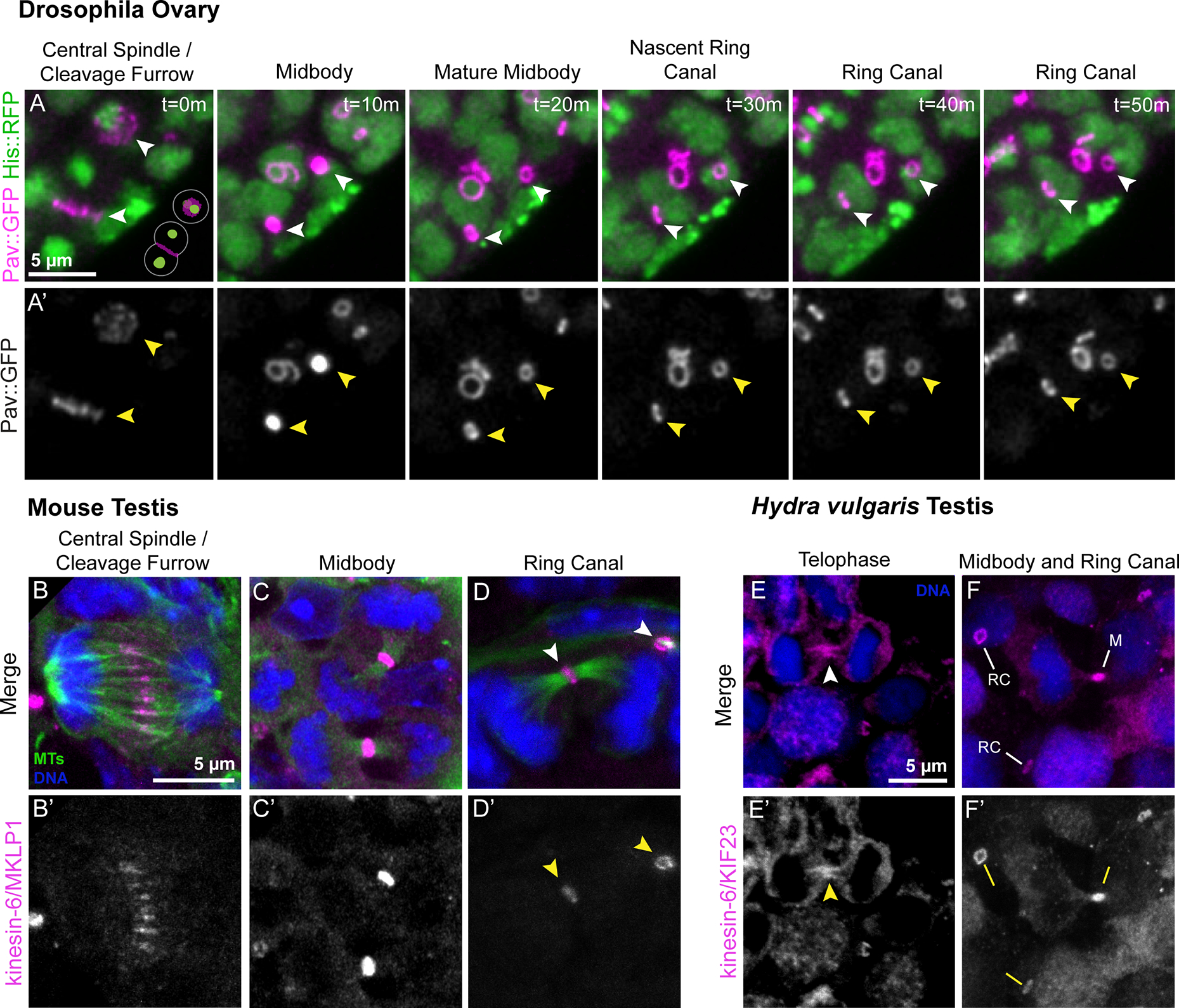
(A) Stills from a time-lapse movie of a germarium expressing Pav::GFP and Histone::RFP. In the plane of focus are two actively dividing cells of a four-cell cyst; the planes of division are oriented perpendicular to each other as depicted in the schematic at the lower right of t=0m. Arrowheads indicate the progression of cleavage furrows to ring canals. B) Mouse testis sections were fixed and stained to detect microtubules (MTs, green), MKLP1 (magenta), and DNA (blue). A total of 242 cells across 11 preparations were analyzed. (B-B’) A dividing germ cell at anaphase. (C-C’) MKLP1-labeled midbodies are present at the intercellular bridge between segregated sister chromatids. (D-D’) Nascent ring canals (arrowheads) are marked by the association of open ring canal with luminal microtubules. (E-F’) Hydra testes were fixed and stained to detect KIF23 (magenta) and DNA (blue). A total of 572 cells across five preparations were analyzed. (E-E’) A dividing spermatocyte with enrichment of KIF23 at the intercellular bridge (arrowhead). (F-F’) Ring canals (“RC”) and a midbody (“M”) are present in the field of view.
Next we investigated whether the reorganization of germ cell midbodies represents an evolutionarily conserved feature of ring canal biogenesis by examining ring canal formation in the male germlines of distantly related animal species, mice and Hydra. There is evidence suggesting that midbodies are involved with ring canal formation in mice; examination of spermatogenesis in tex14−/− mutant mice revealed the presence of MKLP1-labeled midbody core-like foci17. However, it was not clear whether midbodies are a normal step in the ring canal pathway or the consequence of failure to inhibit abscission caused by lack of TEX14. Therefore, we carefully examined the localization of MKLP1 (Pav ortholog) and tubulin at the mitotic intercellular bridges in testis sections from postnatal day 8 mice (Figure 3B–D’). 21.2% of MKLP1 signal was at anaphase spindles (n=51/242; Figure 3B–B’) and the majority of MKLP1 signal was found at ring canals (76.4% (n=185/242); Figure 3D–D’), as expected. However, we also observed MKLP1-labeled midbodies in dividing spermatogonia (2.4%, n=6/242 of all MKLP1-positive structures; Figure 3C), providing evidence that germline midbodies form during mouse cell divisions leading to ring canals. Additionally, we found that the levels of MKLP1 appeared to decline as ring canals formed (compare Figure 3C’ to 3D’), as we observed in Drosophila. These data confirm the presence of midbodies in dividing germ cells of the mouse testis and support the conclusion that a midbody-to-ring canal transition is conserved among bilaterians.
To determine if this germ cell midbody behavior is conserved outside of bilaterians, we examined the localization of the MKLP1 ortholog KIF23 in testes of the cnidarian Hydra vulgaris. Hydra spermatogenesis takes place in an anatomically simple testis that does not have additional accessory structures as found in more recently evolved metazoans30. Hydra ring canals have been documented in electron microscopy studies4, but the identity of ring canal proteins was unknown. Examination of single-cell sequencing datasets revealed that kif23 transcripts were enriched in male germ cell and interstitial cell lineages31, which are cell types known to have intercellular bridges. To study KIF23 localization, we generated and validated a polyclonal antibody against Hydra KIF23 amino acids 208–686 (Figure S5), a region homologous to the one previously used to generate an antibody to Drosophila Pav21. We stained Hydra testes with anti-KIF23 (Figure 3E–F’) and found that 1.7% (n=10/572) of KIF23 was present at the central spindle between anaphase chromosomes, 3.2% of KIF23-labeled structures (n=18/572; Figure 3E–E’) were germline midbodies that localized to the cytoplasmic bridge connecting dividing cells, and the majority of remaining KIF23 localized to mature ring canals (n=544/572; Figure 3F–F’).
From these data, we conclude that the formation of ring canals from germline midbodies is an evolutionarily conserved process and that the midbody-to-ring canal transition represents a key step in ring canal biogenesis.
Midbody reorganization and stabilization of the midbody ring facilitates ring canal formation
To further characterize the germline midbody-to-ring canal transition, we performed live imaging of other cytokinesis proteins during ring canal formation: Tumbleweed/Tum (the binding partner of Pav; sqhP-sfGFP::tum), Anillin (nos>uasp-mRFP::anillin), Myosin-II (sqhP-sqh::GFP), Septin2 (sep2P-sep2::gfp, as a representative Septin subunit), and Sticky (Citron kinase, sqhP-sfGFP::sticky) (Figure 4). GFP::Tum localized similarly to Pav during ring canal formation revealing that both components of the centralspindlin complex were in midbodies that reorganized into ring canals (Figure 4A–A’).
Figure 4. Midbody reorganization and stabilization of the midbody ring facilitates ring canal formation.
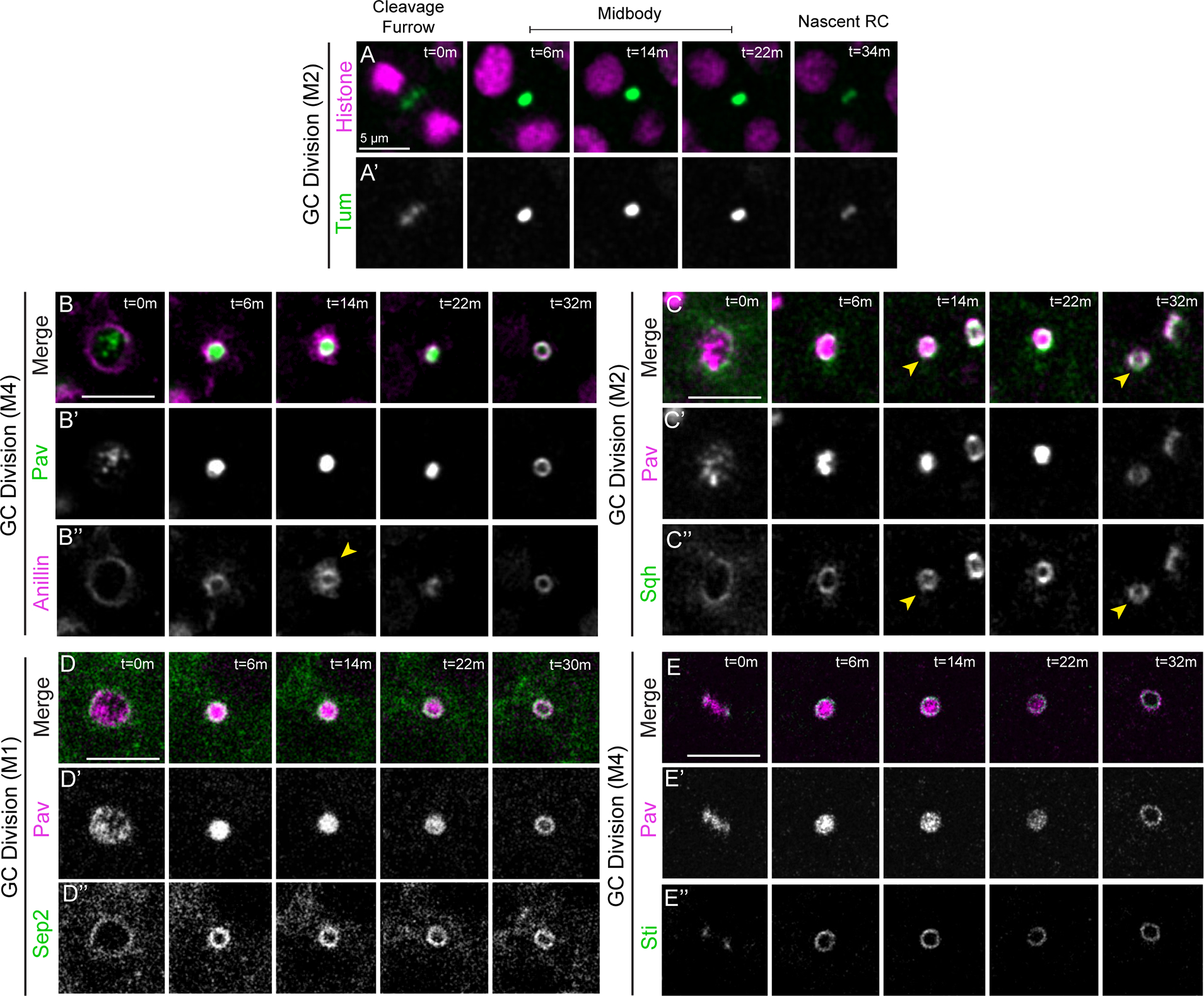
Live imaging of ring canal and cytokinesis proteins during ring canal formation in the Drosophila testis. The mitotic division corresponding to each ring canal is labeled to the left of each montage. (A-A’) GFP::Tum and His::mRFP (n=13). (B-B”) mRFP::Anillin and Pav::GFP (n=22). The yellow arrowhead marks sites of apparent membrane shedding characteristic of midbody ring maturation. (C-C”) Pav::mCherry and Sqh::GFP (n=33). The yellow arrowhead marks the ring canal being monitored over time. (D-D”) Pav::mCherry and Sep2::GFP (n=16). (EE”) Pav::mCherry and GFP::Sticky (n=15). (A-E) m, min. Each panel is a single Z plane. GC, germ cell; M1–4, mitotic division 1–4. Images taken every 2 minutes.
We examined whether the newly formed centralspindlin ring co-localized with proteins in the pre-existing midbody ring. mRFP::Anillin localized to the cleavage furrow and Pav::GFP was mainly detected in the spindle midzone at this stage (Figure 4B–B”). Following contractile ring constriction, mRFP::Anillin localized in a ring around the Pav::GFP-labeled midbody. The mRFP::Anillin appeared to undergo “shedding” (Figure 4B”, t=14 minutes), a term used to describe the membrane pruning that occurs during transformation of the contractile ring to midbody ring in Drosophila S2 cells32–34. Following germ cell midbody reorganization, Pav::GFP and mRFP::Anillin were co-localized in the nascent ring canal (Figure 4B, t=32 minutes). Similarly, Sqh::GFP appeared first as a ring around and then co-localized with the Pav::mCherry signal during ring canal formation and was a robust marker of mature male ring canals as has been described35 (Figure 4C–C”). Sep2::GFP localized to the cleavage furrow with Pav::mCherry and then formed a ring around the midbody (Figure 4D–D”). We also observed that the midbody ring organizing protein Sticky localized in a ring around the Pav focus and then co-localized with Pav after the midbody-to-ring transition (Figure 4E–E”). These data reveal that the centralspindlin ring resulting from the midbody-to-ring reorganization co-localizes with all four midbody ring proteins we examined, suggesting centralspindlin presence could stabilize the midbody ring to form a ring canal.
The midbody organizing and stabilizing protein Citron kinase/Sticky localizes to germline midbodies and nascent ring canals
Given the probable link between midbody rings and ring canals, we further examined the behavior of the midbody organizing protein Citron kinase (Citk), known as Sticky in Drosophila, during ring canal formation. Citk plays a significant role in the establishment and maintenance of proper midbody core and midbody ring architecture during late cytokinesis in cultured cells36. Citk is also indirectly required for cytokinetic abscission through its role in scaffolding and regulating a network of contractile ring components and microtubule-associated proteins10,36,37. Interestingly, mutations in citk cause male sterility in mice due to the lack of intercellular germline bridges as a consequence of complete cytokinesis38,39. In Drosophila spermatogenesis, mutations in sticky result in an increase in multi-nucleate spermatocytes due to failed cytokinesis as well as smaller than average ring canals, attributed to over-constriction of the cleavage furrow38.
To confirm the presence of Sticky at ring canals that we saw with overexpression of transgenic Sticky (Figure 4E), we used antibody staining of fixed tissue to examine Sticky localization in mitotic and post-mitotic cells during spermatogenesis. Sticky antibody staining showed localization at the midbody ring and nascent ring canals in spermatogonia but was undetectable at mature ring canals in spermatocytes (Figure 5A–G). We also observed cytoplasmic punctate structures, presumed to be aggregates, as described for mammalian Citron kinase (Figure 5C”)40,41. While transgenic GFP::Sticky was detectable at the fusome, a branched organelle that extends through all ring canals in germline cysts (Figure S6), we could not detect Sticky at fusomes in our immunostaining experiments, suggesting that Sticky either ectopically localizes to fusomes when over-expressed or epitope masking and inaccessibility of the antibody precludes our ability to visualize Sticky at these structures. Notwithstanding these caveats, these data suggest that transient localization of Sticky may facilitate the midbody-to-ring canal transition.
Figure 5. The midbody organizing and stabilizing protein Citron kinase/Sticky localizes to germline midbodies and nascent ring canals and facilitates the midbody-to-ring canal transition.
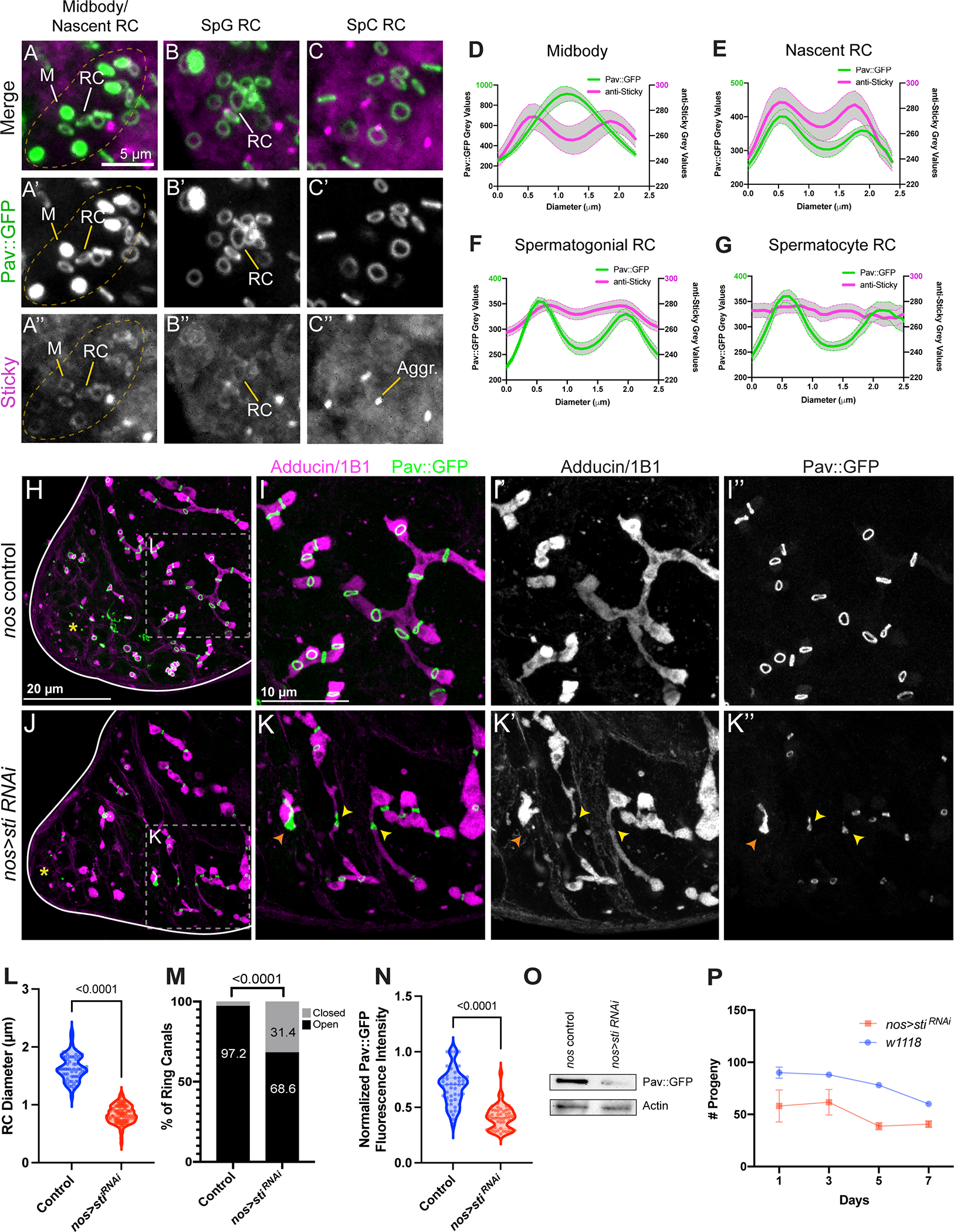
(A-C”) Control Pav::GFP testes were fixed and stained for Sticky protein with a polyclonal anti-Sticky antibody (n=30). (A-A”) An actively dividing 8-CC, with Pav-labeled midbody foci. M, midbody focus; RC, ring canal. (B-B”) A spermatogonial (SpG) cyst with older ring canals. (C-C”) A cyst of primary spermatocytes (SpC). (D-G) Quantification of Pav and Sticky fluorescence intensities from the respective ring canal stages in (A-C). Pav::GFP fluorescence intensity (green trace, left Y axis) versus Sticky (magenta trace, right Y axis) is plotted; midbody (n=9), nascent ring canal (n=5), spermatogonial ring canals (n=20) and spermatocyte ring canals (n=13). Thick lines are the means and error bars (shaded grey) reflect the SEM. (H-I”) Control Pav::GFP testes were fixed and stained with Adducin/1B1 (magenta). The apical tip is marked by a yellow asterisk. (J-K”) nos>sti RNAi testes (n=30). Yellow arrowheads mark closed ring canals and points where fusome structure is perturbed. (L) In nos>sti RNAi testes where the ring canal diameter can be scored (control n=52; nos>sti RNAi n=54), ring canal diameter is reduced. (M) nos>sti RNAi testes have a higher percentage of foci (“closed”) (n=116/369) as compared to controls (n=253/369) (p=<0.0001, fisher’s exact test). (N) Pixel intensities of Pav::GFP are reduced in nos>sti RNAi testes. (O) nos>sti RNAi testes have less total Pav::GFP via Western blot (n=2 biological replicates). (P) nos>sti RNAi animals are sub-fertile as compared to controls (p=0.0063, paired two-tailed t-test).
Perturbation of midbody ring formation and/or stability prevents ring canal formation
If ring canals represent persistent midbody rings, then perturbation of midbody architecture and/or midbody ring stability should result in defects in ring canal formation. The localization of Sticky to germline midbody rings prompted us to examine the effect of sticky knockdown on the midbody-to-ring canal transition. We examined Pav::GFP in testes with sticky expression reduced by RNAi. To provide another metric of proper ring canal formation, we labeled tissues with Hts/Adducin antibodies to visualize the fusome (Figure 5H–I”’, 5J–K”’). In control testes, ring canals were uniform in size with an average diameter of 1.6 μm (±0.02 μm, SEM), with a continuous fusome filling the ring canal lumens (Figure 5H–I”, 5L). In contrast, ring canals in nos>sti RNAi testes were much smaller with an average ring canal diameter of 0.84 μm (±0.02 μm, SEM) (Figure 5J–K”, 5L). Interestingly, we observed that the ring canal phenotype was not restricted to spermatocytes, as previously described by Naim et al.38, but manifested earlier in mitotically dividing cells demonstrating a general requirement for Sticky in ring canal formation during all stages of spermatogenesis (Figure 5J–K”). In addition, 31.4% of Pav::GFP-positive structures were focal with no discernable lumen, appearing as persistent midbodies (Figure 5J–K”, 5M). At these focal sites, the fusome was either discontinuous with obvious gaps or a thread-like structure, indicative of perturbed or complete cytokinesis (yellow arrowheads, Figure 5K–K”). We also observed large aggregates of Pav::GFP protein (Figure 5K), the likely result of ingression of multiple cleavage furrows following failed cytokinesis in the previous cell cycle. Consistent with these defects, we found a reduction in fecundity (Figure 5P), likely due in part to the decline in GSC numbers over time as a result of failed cytokinesis in the GSC-GB division. Our analysis of sticky knockdown testes also revealed a significant decrease in the fluorescence intensity of Pav::GFP in ring canals (Figure 5N) as well as a reduction of total Pav::GFP protein in testis lysates (Figure 5O). The large number of closed ring canals and reduction in Pav fluorescence intensity is consistent with the observed reduction in Pav fluorescence intensity during maturation of the GSC-GB midbody (Figure S2L–L”), suggesting that these closed ring canals are in fact mature midbodies. Together, these data reveal that Sticky is necessary to maintain appropriate levels of Pav in the ring canal and suggest that Pav protein levels in the midbody influence ring canal formation.
To test this idea, we performed live imaging of Pav::GFP in nos>sti RNAi testes (Figure 6A–D). In nos-Gal4 control testes, the time to ring canal formation from the onset of cleavage furrow ingression was an average of 26.7 minutes (± 2.3 minutes, SEM) (Figure 6A, 6E). Of the ring canals that formed successfully in nos>sti RNAi testes, the average time to ring canal formation was highly variable, from 39 to 1260 minutes (Figure 6E). However, many midbodies failed to resolve into ring canals over the course of our time-lapse imaging. We observed that the diameter of the Pav::GFP-labeled midbody directly correlated with whether or not a ring canal formed and also greatly influenced the timing of ring canal formation. For instance, when midbodies were <1 μm in diameter, ring canals failed to form over the course of 12+ hours (Figure 6B, 6F, n=5). However, of midbodies that were similar in size to control midbodies (between 1 and 1.5 μm), ring canal formation was observed but was significantly delayed with times ranging from 40–100 minutes (Figure 6C, 6F, n=8). Interestingly, large midbodies with diameters >1.5 μm that resulted from supernumerary cleavage furrows formed ring canals that were much larger than control ring canals (Figure 6D). Furthermore, the time necessary to resolve these structures was often longer than 9 hours (Figure 6F, n=3).
Figure 6. Perturbation of midbody formation/stability affects ring canal formation.
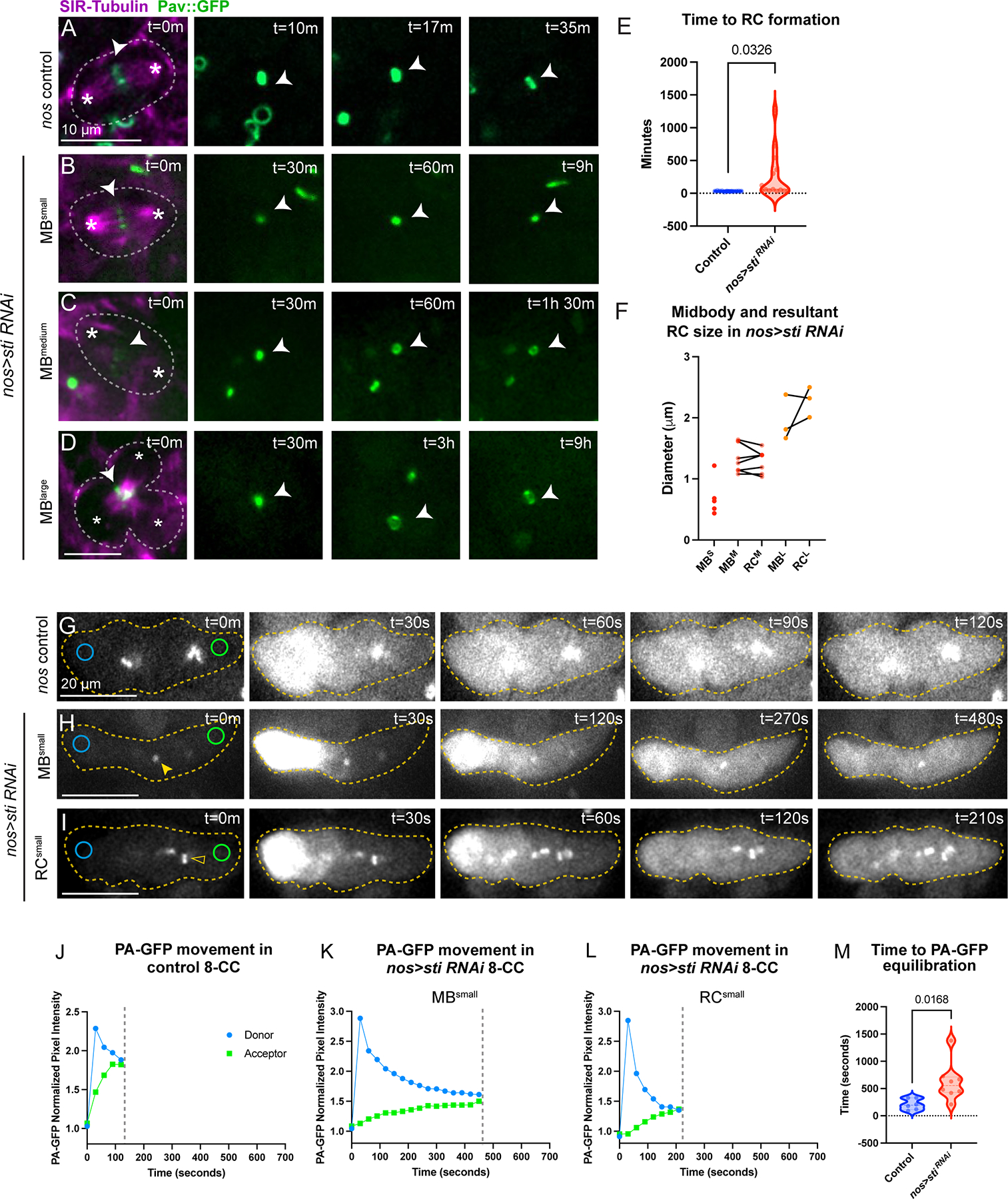
(A) Live imaging of Pav::GFP control testes labeled with SiR-Tubulin to mark microtubules. (B-D) nos>sti RNAi testes with variable midbody (MB) sizes: (B) MBsmall (n=6), (C) MBmedium(n=6), and (D) MBlarge (n=3). Each panel is a single Z plane. Images acquired every 30 minutes. (D) A multipolar spindle (marked with 3 asterisks) and large aggregate of Pav protein (denoted by asterisk) that resulted from failed cytokinesis in the previous cell cycle. (E) The time to ring canal formation is highly variable; the average time to ring canal formation is delayed in nos>sti RNAi testes (n=14, unpaired two-tailed t-test, p=0.0326). (F) Before-and-after plot of midbody and resultant ring canal size in nos>sti RNAi testes. The size of the nos>sti RNAi midbody is directly correlated with whether a ring canal forms as well as the size of the resultant ring canal. (G-I) Live imaging of photoactivatable GFP (PA-GFP) controls (n=6) and nos>sti RNAi (n=7) 8-CC cysts. (H) nos>sti RNAi cyst with small midbodies (closed yellow arrowhead); movement of PA-GFP is much slower than in control cysts. (I) nos>sti RNAi cyst with small, yet open, ring canals (open yellow arrowhead); PA-GFP movement is faster than in cysts where closed ring canals/small midbodies are observed but slower than in controls. Each image is a single Z plane, as such not all cells of the 8-CC are visible in the focal plane. Images acquired every 30 seconds. (J-L) Quantification of PA-GFP movement in the representative montages from (G-I). (M) The average time to PA-GFP equilibration is statistically different between controls and nos>sti RNAi (unpaired two-tailed t-test, p=0.0168).
To understand the functional implications of an untimely or failed midbody-to-ring canal transition, we examined the effect of sticky knockdown on cytoplasmic sharing between germ cells in a cyst. We followed the movement of photoactivatable GFP (PA-GFP) in control and nos>sti RNAi 8-cell cysts (8-CC) in intact testes that were also expressing Pav::GFP to label ring canals (Figure 6G–I). As movement between connected cysts is very rapid in wild-type cysts22, to gain better temporal resolution we photoactivated one cell at the end of an 8-cell cyst (donor cell) and tracked the movement of photoactivated protein to the far end of the cyst (acceptor cell). Movement of PA-GFP between distal cells in control 8-cell cysts occurred quickly (Figure 6G, 6J). The average time to equilibration of PA-GFP (equal PA-GFP pixel intensity values) between the donor and acceptor cells was 210 seconds (±41.7 seconds, SEM; n=6) (Figure 6M). In contrast, movement of PA-GFP in nos>sti RNAi cysts with aberrant midbodies/ring canals was significantly delayed, requiring an average time of 621 seconds (±123 seconds, SEM; n=8) (Figure 6H, 6I, 6K, 6L). The equilibration time in nos>sti RNAi cysts was highly variable (420 to 1380 seconds) and correlated with the size of the midbody/ring canal. Cysts with small midbodies displayed the longest time to equilibration (Figure 6H, 6K) whereas cysts with open, yet small, ring canals displayed equilibration times that were more similar to controls (Figure 6I, 6L).
Together, these data reveal that Sticky plays an important role in the timely resolution of ring canals from germline midbodies via its role as a midbody organizer and stabilizing protein. Failure to transform germ cell midbodies to ring canals in sti RNAi affects cytoplasmic sharing between cells, which could impact the movement of important molecules (cell cycle or other) necessary for the production of functional sperm.
Germ cell midbodies do not initiate abscission
The characteristic absence of abscission during incomplete cytokinesis suggests that the germline midbody may function differently from the midbodies formed during complete cytokinesis. During cytokinesis in completely dividing cells, centralspindlin at the midbody links to the abscission machinery through a series of direct protein-protein interactions. In Drosophila, the ESCRT-associated protein Alix binds Pav directly to recruit the ESCRT-III complex that facilitates abscission42. ESCRT-III complex subunits then recruit the microtubule severing complex Spastin, which promotes disassembly of microtubules in the midbody in preparation for abscission43.
We investigated the localization of abscission pathway proteins involved with membrane abscission and microtubule severing (Figure S7). While we could often detect GFP::Alix (nos>UAS-GFP::alix) at germ cell midbodies in fixed preparations and with live imaging (Figure S7A), similar to its localization to the midbodies during complete cytokinesis in Drosophila female germline stem cells44, we did not detect ESCRT-III/Shrub::GFP (nos>UAS-shrub::GFP) or Spastin::GFP at midbodies or nascent ring canals even when over-expressed (Figure S7B, S7C, respectively). These data suggest that midbody-mediated abscission is blocked early in the process after ALIX recruitment.
Recent research has demonstrated a role for the USP8 de-ubiquitinase in ESCRT-III inhibition during ring canal formation45. To investigate whether USP-8 functions before or after the midbody-to-ring canal transition, we examined Pav::GFP in testes with RNAi knockdown of usp-8. Interestingly, reduced usp-8 expression did not completely block ring canal formation (Figure 7E), suggesting the midbody-to-ring transformation still occurred. Ring canals formed at the apical end of the testis similar to controls (compare Figure 7A–B with 7E–F, white arrowhead). However, as cysts developed, ring canals collapsed into ectopic midbodies, beginning with the older ring canals (Figure 7F, yellow arrowhead), causing ultimate fusome fragmentation as observed by Mathieu et al.45 (compare Figure 7C–D with 7G–H, yellow arrowheads). Importantly, the diameters of the intact ring canals in usp-8 RNAi testes (1.8 μm ± 0.02 SEM, n=86) were similar to control ring canals (1.7 μm ± 0.02 SEM, n=71) (Figure 7I). However, the distribution of ring canals sizes was skewed in nos>usp8 RNAi testes as there were more ring canals of smaller diameter, consistent with the observations of ring canal collapse that precedes formation of ectopic midbody foci made by Mathieu et al.45.
Figure 7. Germ cell midbodies do not initiate abscission.
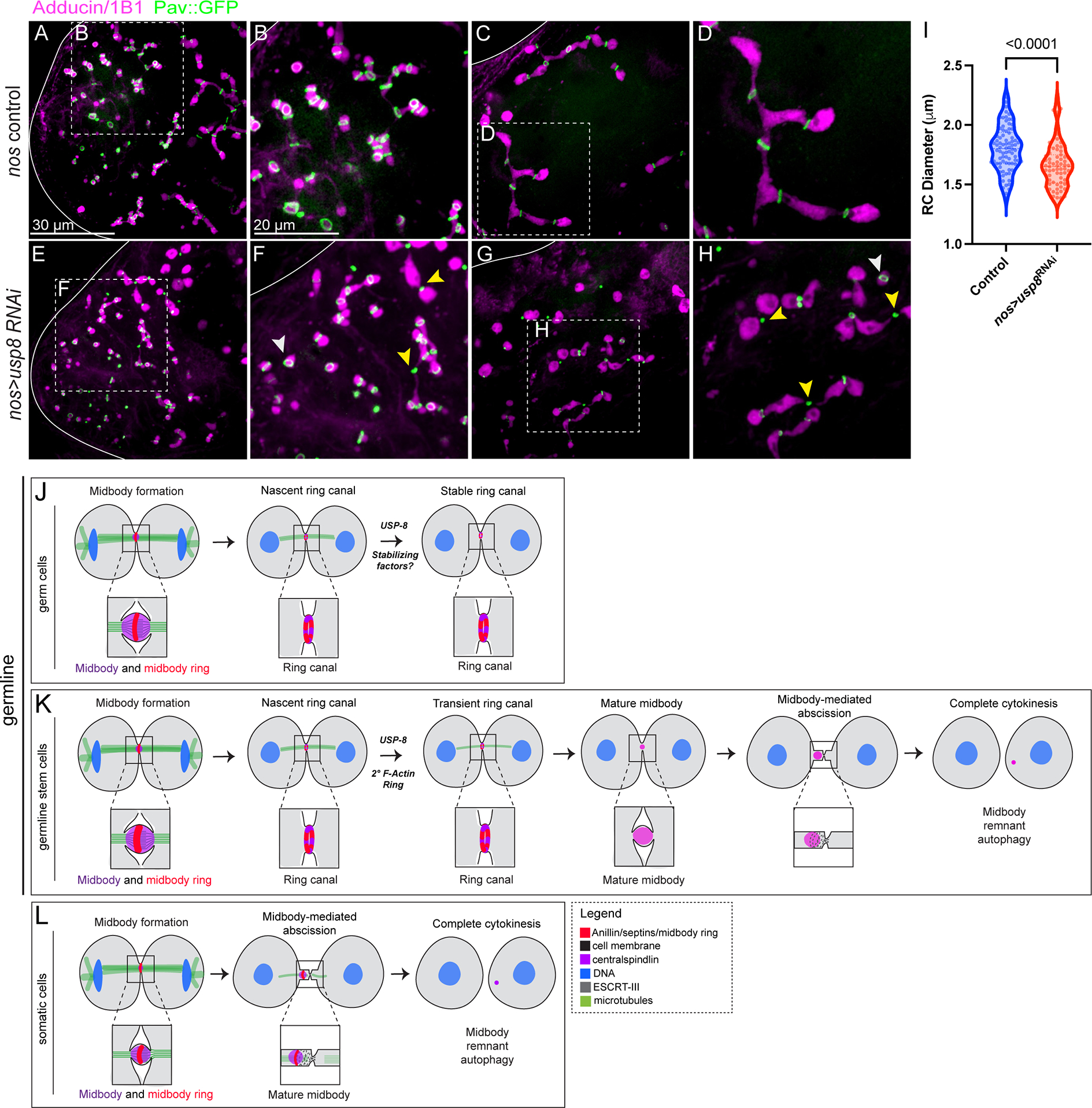
(A) Pav::GFP-expressing control testes were fixed and stained with Adducin/1B1 to visualize the fusome (n=40). (A-B) The apical end of the testis. (C-D) The primary spermatocyte region of the testis. (E-H) nos>usp8 RNAi testes fixed and stained with Adducin/1B1 (n=36). (E-F) Ring canals (white arrowheads) form in testes depleted of USP-8, but ectopic midbodies (yellow arrowheads) are observed and the fusome remains intact. These midbodies correspond to the oldest ring canals formed during the first mitotic division, consistent with Mathieu et al.45. (G-H) In the primary spermatocyte region, many ring canals have collapsed to form ectopic midbody foci and the architecture of the fusome is consistent with fragmentation as a result of complete cytokinesis. (I) Quantification of ring canal diameters in control (n=86) and nos>usp8 RNAi (n=71) testes. Violin plots reveal that the range of ring canal sizes in unchanged in nos>usp8 RNAi testes, but the distribution of ring canal sizes is altered (unpaired two-tailed t-test, p<0.0001). Ring canals of smaller diameter are more prevalent in nos>usp8 RNAi testes consistent with ring canal collapse. (J-L) Diagram of midbody behavior during cell division in the three cell types in the Drosophila testis that we examined. (J-K) Midbody reorganization to form a ring canal is observed in both the germline stem cells and the germ cells, but absent from the somatic and completely dividing cyst stem cells (depicted in L).
Taken together, these data suggest USP-8 is not strictly required for ring canal formation, and ring canal formation precedes abscission inhibition.
Discussion
Until now, studies of ring canal formation have lacked the spatiotemporal resolution required to understand the pathway of ring canal biogenesis. Our results reveal the temporal dynamics of incomplete cytokinesis and ring canal formation for the first time and we demonstrate that germ cell ring canal formation occurs via an evolutionarily conserved midbody intermediate.
Our live imaging of dividing germline cells in the Drosophila testis and ovary challenge previously held ideas of contractile ring arrest, and inform a model that is consistent with observations of germ cell midbodies during spermatogenesis in mice (Figure 7I). Following midbody formation in dividing germ cells, the midbody core reorganizes to join proteins in the midbody ring, which is then stabilized to form a ring canal (Figure 7I). Our data reveal a role for the evolutionarily conserved centralspindlin complex in ring canal formation, as it undergoes a stereotypic re-localization that results in ring canal lumen formation. The centralspindlin complex joins Anillin, Sep2, Sqh/NMY-II, and Sti/Citron kinase in the midbody ring to form the nascent ring canal. We also demonstrate a role for the conserved midbody organizing protein Sti in controlling the size of ring canals, the timing of ring canal formation, and whether they form at all. Finally, we show that ring canals form before USP-8-mediated inhibition of abscission, revealing the possibility that midbody remodeling is separate from abscission inhibition. Incomplete cytokinesis is therefore the likely result of two coordinated pathways: midbody reorganization and abscission inhibition.
Germ cell midbody formation and subsequent remodeling is an intrinsic feature of germline cells during gametogenesis
Our investigation of dividing germline cells in distantly related species reveals a mechanism of incomplete cytokinesis giving rise to ring canals that is conserved between the sexes and across evolution. Surprisingly, through our characterization of ring canal formation, we discovered that incomplete cytokinesis shares more features with complete cytokinesis than previously appreciated. Rather than the pathways of cytokinesis diverging shortly after cleavage furrow formation and ingression, our data suggest that both cytokinesis programs assemble a midbody that appears to act as a pause point, allowing the midbodies to either recruit abscission components that completely separate newly formed daughter cells (Figure 7K), or to quickly reorganize to form a ring canal that prevents abscission and leads to the formation of interconnected cells in a syncytium (Figure 7I).
We were surprised to discover that germline stem cells also undergo an initial midbody-to-ring transformation considering that they ultimately undergo complete cytokinesis. However, the resulting intercellular bridges are transient; after several hours they collapse into a second much smaller midbody that mediates abscission (Figure 7J). Therefore it appears that midbody reorganization may be a common feature of all dividing germ cells. The significantly longer cell cycle time for germline stem cell division compared to germ cell divisions may allow time for abscission proteins to accumulate at the transient ring canals and drive complete cell division, as suggested by Mathieu et al.45.
The midbodies of dividing somatic cyst cells do not appear to undergo a midbody-to-ring transition. This clear difference between germline and somatic cells in the testis underscores the possibility that midbody remodeling is intrinsic to germline cells. However, in some immortalized human somatic cells in culture, MKLP1 has been detected in both midbody cores and midbody rings, suggesting the possibility of a midbody-to-ring transformation23. As abscission can take several hours in some cultured cells36, it will be interesting to more closely examine with live imaging whether these cells also undergo a similar midbody-to-ring transition – this would suggest that transient ring canal formation is a general phenomenon of delayed abscission rather than being specific to germline cells. Live imaging of midbodies in additional tissue-intact dividing somatic cells is needed to determine whether midbody remodeling is part of normal somatic cell division.
Mechanistic insights into midbody remodeling and midbody ring stabilization
The reorganization of centralspindlin in the midbody core to join proteins in the midbody ring (Figure 4) suggests that ring canals originate from stabilized midbody rings. There are several possible mechanisms for midbody remodeling for future research. As centralspindlin reorganizes from a midbody core to a ring, protein in the center of the focus disappears, either via targeted degradation or relocalization, leaving behind a centralspindlin ring that colocalizes with Sticky, Anillin, NMY-II, and the Septins. Perhaps in the same way that Sticky retains Anillin during the contractile ring to midbody ring transition in Drosophila S2 cells33,34, Sticky could contribute to the selective retention of centralspindlin protein at the membrane during ring canal formation.
Another possibility is that specific microtubule interactions may facilitate ring canal formation. During the midbody to ring transition, the microtubule motor domain of MKLP1 could release the microtubules of the spindle midzone and, in doing so, MKLP1 reorganizes to form a ring that encircles the microtubule bundle. In support of this model, studies in Hela cells demonstrate that the microtubule motor domain of MKLP1 is not required for localization to the midbody ring23. Alternatively, there may be two sub-populations of centralspindlin within the midbody: i) the cortically associated ring population that is protected from degradation, and ii) the spindle midzone associated population that is selectively degraded during the midbody-to-midbody ring/ring canal transition.
Finally, Drosophila ring canals are rich in phosphotyrosine epitopes and glycoproteins1,8,46, but whether these post-translational modifications are involved with producing or retaining ring canals is an area for future research.
Citron kinase contributes to germ cell midbody formation
Our research revealed that the serine/threonine kinase Citron kinase/Sticky plays an important role in ring canal biogenesis. Our observations of Sticky function during ring canal formation are similar to descriptions of the Sticky-mediated contractile ring-to-midbody ring transition that occurs during cytokinesis in somatic Drosophila S2 cells33,34. In these cells, Sticky is necessary to stabilize the constricting contractile ring and to retain Anillin at the mature midbody ring34. Depletion of Sticky disrupts the organization of the midbody and the stability of the associated midbody ring resulting in either binucleation caused by cytokinesis failure prior to abscission or by premature abscission34,47. The ectopic midbodies and associated breaks in the fusome that form in sticky RNAi testes suggest that complete cytokinesis occurs at these sites, consistent with premature abscission in S2 cells34,47. Therefore, it appears that Sticky function in somatic midbody rings and during germline ring canal formation is analogous as Sticky plays important roles in stabilizing the midbody ring that delays or inhibits abscission.
Do ring canals reflect the evolution of multicellularity?
Our findings in distantly related species underline the evolutionary conservation of germline midbody reorganization during incomplete cytokinesis that results in ring canals, which may reveal insight into the evolution of cell division processes. During certain life cycle stages in single-celled colony-forming eukaryotes like the choanoflagellates, the closest living relatives of animals, and in the green algae Volvox, neighboring cells in colonies are connected by intercellular bridges48–50. It is unclear whether midbody proteins localize to the intercellular bridges in the colonies as they do in germline ring canals. However, given the presence of centralspindlin homologs in these genomes, it is exciting to consider that centralspindlin dynamics during incomplete cytokinesis may have contributed to the ancestral program that produced chains of cells during the evolution of multicellularity. Later embellishments in the form of abscission proteins could have produced completely separated cells while the original function of the midbody ring to form a ring canal was lost.
As incomplete cytokinesis events are observed during normal development and are prevalent in disease states, investigation into the mechanism of ring canal formation is poised to reveal insights into the fundamental mechanism of incomplete cytokinesis across biology.
Limitations of the study
Drosophila germ cells are not ideal for an approach using bulk biochemical purification of midbodies to identify putative germ cell-specific midbody components. Thus, we focused on studying known cleavage furrow and midbody proteins using live imaging of tagged proteins in intact tissue. Because ring canals contain proteins that function during cytokinesis, it is difficult to use genetics to pinpoint their function in ring canals. Mutations in pavarotti and sticky cause pleiotropic phenotypes including defective cell division in several cell types. Using germ cell-specific RNAi knockdown of sticky gene expression allowed us to characterize ring canal phenotypes. Further work is needed to decipher the molecular functions of Citron kinase (encoded by sticky) and Pavarotti. Future studies will determine whether Citron kinase functions directly as a scaffolding protein in ring canals or indirectly as a kinase during ring canal formation.
STAR Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lynn Cooley (lynn.cooley@yale.edu).
Materials availability
Data reported in this paper and reagents generated in this study will be shared by the lead contact upon request.
Data and code availability
Plasmids, fly lines, and microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Raw data from Figure S5 was deposited on Mendeley at https://data.mendeley.com/datasets/4bdxw3v44s/draft?a=49439b09-22c3-4da4-a02f-596c371c514a
Any additional information required to reanalyze the data reported is available from the lead contact upon request.
Experimental model and subject details
The experimental models in this study include Drosophila melanogaster, Mus musculus, and Hydra vulgaris. Information on developmental stages used is described in the methods (see below). All fly lines were kept in mite-free conditions and grown at room temperature in the lab in plastic vials or bottles supplemented with fly food (1.8% m/v yeast, 1% m/v soy flour, 7.3% m/v cornmeal, 0.6% m/v agar, 7.6% v/v light corn syrup, 0.48% v/v propionic acid). All fly lines were flipped to new vials or bottles every two weeks. Hydra were kept at 18°C in a temperature-controlled incubator and fed 2–3 times per week. All mouse procedures were performed under the approval of Yale University’s Institutional Animal Care and Use Committee. CD1 male mice were used in this study and testes were dissected at post-natal day 8 in accordance with approved animal protocols. Mice were maintained in a barrier facility with 12-hour light/dark cycles.
Fly husbandry and strains
The following fly strains were obtained from the Bloomington Drosophila Stock Center: nos-Gal4 (#7303), his2av::mRFP (#23651), his2av::eGFP (#24163), sti shRNA (#35392), sep2::GFP (#26257), sfGFP::sti (#81646), sqh::GFP (#57145), mRFP::anillin (#52220), shrub::GFP (#32559) and sfGFP::tum (#76264). We imaged Pavarotti (Pav) at endogenous levels with Pav::GFP22 or with Pav::mCherry (this study). uasp-GFP-alix44 flies were obtained from Kaisa Haglund (Oslo University Hospital) and uas-spas-eGFP51 flies from Damien Brünner (University of Zurich). 20XUAS-mC3PA-GFP was provided by Pfeiffer et al.52. We examined the behavior of microtubules with a GFP-tagged microtubule-binding protein Jupiter::GFP53,54 using the Jupiter::GFP FlyTrap line (CB05190).
Method details
Construction of Pav::mCherry
Pav::mCherry was generated in a w1118 background by replacing the endogenous stop codon of pav with a C-terminal mCherry-LoxP-3XP3-eGFP-LoxP knock-in cassette via CRISPR/Cas9-mediated genome editing by homology-dependent repair (performed by WellGenetics, Inc.). Following screening, 3XP3-GFP was removed by crossing to flies expressing Cre recombinase.
Construction of Pav::Dendra2
The coding sequence for Dendra2 was amplified from genomic DNA of ɑ-tubulin 84B::Dendra2 flies (#51315). Full-length Pav cDNA and Dendra2 were cloned into the pUASz vector55 with Gibson Assembly (NEB). Pav fused to Dendra2 is spaced with the flexible linker GGSGGSGGSG. The final plasmid was inserted at the attP40 site on 2L by Genetivision (Houston, TX).
Live imaging in Drosophila testes
Live imaging of ring canal formation in Drosophila testes was performed as previously described35. Newly eclosed males were dissected in Ringer’s solution (200 mM NaCl, 4 mM KCl, 0.3 mM CaCl2 2H2O, 5 mM NaHCO3, 0.13 mM NaH2PO4 H20) and then transferred in Ringer’s solution to a Poly-L-Lysine-coated coverslip bottom of a round imaging dish (MatTek). Ringer’s solution was removed, allowing the testes to settle on the coverslip, and replaced with imaging medium (15% Fetal Bovine Serum, 0.5X penicillin/streptomycin, 0.2 mg/ml insulin in Schneider’s insect medium). Testes were imaged on either a Bruker Opterra II Swept Field, Zeiss 980 laser scanning confocal microscope, Leica Stellaris 8 laser scanning confocal microscope, or Zeiss Axio observer 7 inverted microscope with CrEST X-light v2 spinning disk system and Photometrics Bsi sCMOS camera. Images were acquired with a Z step size of 1 μm using either a 60X water immersion objective lens (Bruker Opterra II, 1.3 NA), W Plan-Apochromat 40X dipping lens (Zeiss, 1.0 NA), HC PL APO CS2 40X oil immersion objective lens (Leica, 1.3 NA), or LD C-Apochromat W Korr 63X water immersion objective lens (Zeiss, 1.15 NA). Images were collected at either 1 minute, 2 minute, or 25 minute intervals. The average Z-distance traveled was 47 μm per time point. Figures were created from, and analyses were performed on either individual Z sections or 2–4 μm Z plane projections. When possible, samples were imaged in a temperature-controlled chamber set to 25°C to prevent thermal drift. For each listed genotype, imaging was performed on at least three separate days, imaging experiments, and from independent crosses. We analyzed the following numbers of movies (each capturing at least one cell/cyst division) for the listed genotypes to generate the data presented: Sqh::GFP, 40; mRFP::Anillin, 33; Septin-2::GFP, 16; Jupiter::GFP, 28; Tum::GFP, 13; Pav::mCherry and Pav::GFP, >100; Pav::Dendra2, 10; GFP::Sti, 15; nos>sti RNAi, 20; nos driving both sti RNAi and PA-GFP, 8; nos>PA-GFP, 6.
Live imaging of microtubules in nos>sti RNAi testes was performed by labeling with SiR-Tubulin (Cytoskeleton, Inc.). SiR-Tubulin was added at a concentration of 100 nM to imaging medium and testes were incubated for one hour before imaging.
Quantification of Pav fluorescence intensities, midbody/ring canal diameters during ring canal formation, timing of ring canal formation, and photoactivatable::GFP
Grey values in a 2 μm2 region of interest corresponding to the nascent midbody and ring canal were measured in Fiji/ImageJ. Measurements were made every five minutes following cleavage furrow initiation and normalized to the fluorescence intensity in the nascent midbody. Consequently, the normalized pixel values are highest (1.0) in the nascent midbody. For measurements of Pav fluorescence intensity in non-actively dividing cells, pixel values were normalized to the first time point to assess photobleaching.
Diameters of midbody foci were quantified from full width at half maximum (FWHM) measurements from line scans obtained in FIJI/ImageJ. XY coordinates were exported to Qtiplot to calculate FWHM. Ring canal diameters were obtained by calculating the distance between the two highest pixel values from a line scan.
Line scan analyses were performed in Fiji/ImageJ by placing a line of 5-pixel width across the center of the ring canal or midbody intermediate. For pixel intensity values, resultant values were normalized to the pixel values of Pav in the midbody focus.
To quantify pixel values of Pav::GFP or Pav::mCherry in fixed tissues, we measured the fluorescence intensity in a 2 μm2 region of interest corresponding to the midbody and ring canal and background intensity was subtracted from each value and the ratio was reported.
To determine the timing of ring canal formation, all times reported are relative to the onset of cleavage furrow ingression.
Quantification of photoactivation was performed in Fiji/ImageJ where a 10 μm2 region of interest was placed in either the donor or acceptor cell or a non-photoactivated neighboring cell. The mean pixel intensity values of the regions of interest in a single Z-slice were measured using the histogram function. The fluorescence intensity of the donor/acceptor cells were then normalized to the pixel values of the non-photoactivated neighbor cell at each time point.
Live imaging experiments were repeated on at least three separate days and a minimum of 6 testes across experiments were analyzed for each genotype.
Photoconversion of nos>pav::dendra2
Testes from nos>pav::dendra2 flies were dissected in Ringer’s solution and prepared for live imaging as described above. Photoconversion experiments were carried out on a Bruker Opterra II Swept Field confocal microscope with PlanApo 40X water immersion objective with 1.6X optical zoom (1.15 NA). A single Z-stack was captured prior to photoconversion. A 10 μm2 region of interest in a single Z-plane was photoconverted with 50 iterations of 405 nm wavelength, a 50 μs pixel dwell time, and 390 nm pixel size (512 × 512 pixels). Following photoconversion, a Z-stack was acquired every 60 seconds. Green-to-red photoconversion was detected using the same settings. Photoconversion experiments were performed on three separate days to obtain at least 10 biological replicates for further analysis.
Photoactivation of photoactivatable::GFP (PA-GFP)
Live testes expressing Pav::GFP and photoactivatable-GFP (PA-GFP) with or without sti shRNA driven by nos-Gal4 were dissected in Ringer’s solution and prepared for live imaging as described above. A 10 μm2 region of interest was photoconverted throughout the Z-dimension of an eight cell cyst with step sizes of 1 micron resulting in an average Z-dimension of 12 microns. Photoactivation was carried out using a 405 nm laser and the average pixel dwell time was 10 μs. Photoactivation was performed on a Bruker Opterra II Swept Field confocal microscope with PlanApo 60X water immersion objective with 1.6X optical zoom (1.2 NA). Activated GFP was observed by capturing the Z-stack using 488 nm excitation every 30 seconds for 5–25 minutes. Four independent photoactivation experiments were performed for each genotype. However, due to the variable nature of photoactivation in intact animals, data were excluded when the photoactivated region drifted from the coverslip or if the region of photoactivation resulted in photoactivation of neighboring cysts that would otherwise make it difficult to discern the migration of GFP protein.
Live imaging in Drosophila ovaries
Live imaging of ring canal formation in Drosophila ovaries was performed as previously described 56. Briefly, newly eclosed females were dissected in Ringer’s solution on a Poly-L-Lysine-coated coverslip of a MatTek imaging dish. Ringer’s solution was removed carefully as to not disrupt the germaria and replaced with imaging medium (15% Fetal Bovine Serum, 0.5X penicillin/streptomycin, 0.2 mg/ml insulin in Schneider’s insect medium). Images were acquired with a Z step size of 0.6 μm using a W Plan-Apochromat 40X dipping lens (Zeiss, 1.0 NA) every 10 minutes. Four separate imaging experiments from independent crosses were performed over a period of four days.
Drosophila immunocytochemistry in testis
Testes from newly eclosed males were dissected in Ringer’s solution and fixed for 10 minutes in 4% paraformaldehyde in PBT (phosphate-buffered saline with 0.3% Triton X-100 and 0.5% BSA). Fixed tissue was washed in PBT and permeabilized in 1% Triton X-100 in PBS for 30 minutes at room temperature. Testes were incubated in blocking solution (10% normal goat serum in PBT) for 30 minutes at room temperature followed by incubation in primary antibody diluted in blocking solution. The following primary antibodies were used: mouse anti-Hts1B1/Adducin (1:50, Developmental Studies Hybridoma Bank) and rabbit anti-Sticky57 (1:600). Alexa Fluor-conjugated secondary antibodies were used at 1:200 and diluted in blocking solution. Samples were washed in PBT and mounted on slides in ProLong Gold antifade reagent (Invitrogen). DNA was visualized with DAPI (1 μg/ml). Samples were imaged with a Plan-Apochromat 63X 1.4 NA oil immersion objective on Zeiss 980 laser scanning confocal microscope with Airyscan2. Airyscan2 images were processed post-acquisition with the Zeiss Airyscan2 processing feature. For each genotype, a minimum of 10 testes from each fixed preparation were analyzed. Each immunolabeling experiment was performed at least three times.
Male fertility tests
Fecundity of sti-depleted males was assessed by pairing a single male with three w1118 virgin females. Males were shifted every two days to new vials containing new virgin females for 14 days. The total number of adult progeny was counted to determine fecundity.
Mouse immunocytochemistry
All mouse work was approved by Yale University’s Institutional Animal Care and Use Committee. Mouse testes from postnatal day 8 mice were fixed in 4% paraformaldehyde in PBS overnight at 4°C, sucrose-protected in 30% sucrose in PBS overnight at 4°C, embedded in Optimal Cutting Temperature Compound (Tissue-Tek) prior to freezing, and samples were stored at −80°C. 10-micrometer sections were cut via cryostat. Slides were post-fixed in 4% paraformaldehyde for 8 minutes at room temperature. Sections were washed briefly in PBS and then incubated in a 1% SDS solution for 5 minutes at room temperature in a humid chamber. Sections were washed thoroughly in PBS and then incubated in blocking buffer (3% BSA, 5% Normal Goat Serum, 5% Normal Donkey Serum in PBS-T (PBS containing 0.2% Triton X-100)) for 30 minutes at room temperature. Primary antibodies were diluted in blocking buffer and incubated in a humid chamber for 30 minutes. The following primary antibodies were used: 1:200 rabbit anti-MKLP1 (clone 7C9, Bioss Antibodies, cat. # BSM-52401R) and 1:10 mouse anti-alpha Tubulin (clone 4A1, Developmental Studies Hybridoma Bank). After washing in PBS-T, sections were incubated in secondary antibody diluted in blocking buffer for 30 minutes. Alexa Fluor-conjugated antibodies were diluted 1:200 in blocking buffer and incubations were done for 30 minutes at room temperature in a humid chamber. DNA was visualized with DAPI (1 μg/ml). Sections were washed and then mounted in ProLong Gold antifade reagent (Invitrogen). Coverslips were sealed with clear nail polish. Tissue sections were imaged on a Zeiss 980 laser scanning confocal microscope with 63X 1.3 NA oil immersion objective.
Hydra culturing
Hydra vulgaris AEP were asexually cultured at 18°C in Hydra medium by standard procedures58 and fed Artemia nauplii 2–3 times a week. Polyps with testes were selected for immunostaining.
Hydra immunocytochemistry
Hydra were relaxed in 2% urethane in Hydra medium for 2 minutes, fixed for 10 minutes in 4% paraformaldehyde in Hydra medium at room temperature, washed 3 times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 15 minutes. Samples were then incubated in blocking buffer (1% BSA, 10% Normal Goat Serum, 0.1% Triton X-100 in PBS) for one hour. Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. Rabbit Anti-KIF23 antibody (this study) was used at 1:200. Samples were washed 3 times in PBS and incubated in secondary antibody diluted in blocking buffer for 2 hours at room temperature. Alexa Fluor-conjugated secondary antibodies were used at 1:200. DNA was visualized with 1 μg/ml DAPI. Animals were mounted in ProLong Gold antifade reagent (Invitrogen). Whole mount Hydra preparations were imaged on a Zeiss 980 laser scanning confocal microscope with 63X 1.3 NA oil immersion objective.
Hydra KIF23 antibody generation and validation
A fragment of Hydra KIF23 (Uniprot T2MND8, GenBank accession number XM_012700539.1) from amino acids 208–686 was cloned into the pET30a (+) expression vector. Recombinant protein was expressed in BL21 Star™ DE3 cells. Purified proteins were used to raise antisera in rabbits (Genscript) from which antibody was then affinity purified. Antibody was verified via Western blot against purified protein that was used as immunogen as well as Hydra whole cell lysate. 50 ng purified protein, 3 Hydra equivalents, and 10 Drosophila testis equivalents were run on a 4–15% gradient SDS-PAGE gel (BioRad) and transferred at 300 mAmps for 42 minutes to a nitrocellulose membrane (GE Healthcare). The blot was probed with 1:5000 anti-KIF23 antibody and two replicates were performed.
Quantification and statistical analysis
Raw image files were adjusted for brightness and contrast in Fiji/ImageJ59. Single Z-slices were used for image analyses. Generation of plots and statistical analysis was performed in GraphPad Prism. Either Student’s t tests or ordinary ANOVAs were used for statistical comparisons. In each analysis, p values less than 0.05 were considered to be statistically significant. Statistical tests used are listed in the figures, figure legends, and the text, where appropriate. With all statistical averages, the standard error of the mean is also presented.
Supplementary Material
Ring canal formation in the Drosophila testis occurs via a midbody-like intermediate. Related to Figure 1.
A time-lapse movie of a testis expressing Pav::mCherry (expressed at endogenous levels; magenta) and His::GFP (green). A dividing two-cell cyst is outlined in a white box. The testis is oriented to the left (apical tip) and the hub is evidenced by the accumulation of Pav::mCherry foci. The movie is displayed at a frame rate of 5 frames per second.
Midbody formation, but not reorganization, in dividing cyst stem cells in the Drosophila testis. Related to Figure 1.
A time-lapse movie of a testis expressing Pav::mCherry (expressed at endogenous levels; magenta) and His::GFP (green). A dividing cyst stem cell is marked with a white arrow. The hub is evidenced by the accumulation of Pav::mCherry foci. The movie is displayed at a frame rate of 5 frames per second.
Ring canal formation in the germline stem cell occurs via a midbody-like intermediate. Related to Figure 1.
A time-lapse movie of a testis expressing Pav::GFP (expressed at endogenous levels; green) and mRFP::Anillin (driven by nos-Gal4, magenta). A dividing germline stem cell is marked with a white arrow. The testis is oriented to the left (apical tip) and the hub is evidenced by the accumulation of mRFP::Anillin foci. The movie is displayed at a frame rate of 5 frames per second. The movie captures the dynamics of Pav and Anilin from the formation of a cleavage furrow to the beginning of collapse of the mature midbody.
Ring canal formation in the Drosophila ovary occurs via a midbody-like intermediate. Related to Figure 3.
A time-lapse movie of a germarium expressing Pav::mCherry (expressed at endogenous levels; magenta) and His::GFP (green). The anterior end of the germarium is oriented down and left. Top panel: merge of Pav::GFP (magenta) and His::RFP (green). Bottom panel: individual channel of Pav::GFP. The frame rate is 5 frames per second.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-MKLP1 antibody (clone 7C9) | Bioss Antibodies | Cat#BSM-52401R |
| Mouse anti-Tubulin, alpha antibody (clone 4A1) | DSHB | RRID: AB_2732839 |
| Rabbit anti-Kinesin-6 polyclonal antibody | This study | N/A |
| Mouse anti-Hts antibody (clone 1B1) | DSHB | RRID: AB_528070 |
| Rabbit anti-sticky polyclonal antibody | D’Avino et al.57 | RRID:AB_2567379 |
| Goat anti-rabbit IgG secondary antibody, AF488 | ThermoFisher | Cat.#A-11008; RRID: AB_143165 |
| Goat anti-mouse IgG secondary antibody, AF568 | ThermoFisher | Cat.#A-11031; RRID:AB_144696 |
| Mouse anti-GFP antibody (clones 7.1 and 13.1) | Roche | Cat.#11814460001; RRID:AB_390913 |
| Mouse anti-Actin antibody (clone JLA20) | Sigma-Aldrich | Cat.#MABT219; RRID:AB_11203498 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ProLong Gold antifade reagent | ThermoFisher | P36934 |
| 8% Paraformaldehyde | Electron Microscopy Sciences | 157-8-100 |
| Poly-L-Lysine | Sigma | P8920 |
| Fetal Bovine Serum | Gibco | A31605-01 |
| Insulin | Sigma | I0516 |
| Penicillin/Streptomycin | Gibco | 15140-122 |
| Schneider’s Insect Medium | Gibco | 15140-122 |
| SiR Tubulin | Cytoskeleton | CY-SC002 |
| Deposited Data | ||
| Figure S5 immunoblot | doi:10.17632/4bdxw3v44s.1 | N/A |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: GAL4::VP16-nos.UTR | Bloomington Drosophila stock center | RRID:BDSC_7303 |
| D. melanogaster: His2av-mRFP | Bloomington Drosophila stock center | RRID:BDSC_23651 |
| D. melanogaster: His2av-eGFP | Bloomington Drosophila stock center | RRID:BDSC_24163 |
| D. melanogaster: UAS-sti RNAi | Bloomington Drosophila stock center | RRID:BDSC_35392 |
| D. melanogaster: Septin2-GFP | Bloomington Drosophila stock center | RRID:BDSC_26257 |
| D. melanogaster: UAS-Spas-eGFP | Jankovics and Brünner51 | N/A |
| D. melanogaster: Pav-GFP | Kaufman and Price et al.21 | N/A |
| D. melanogaster: Pav-mCherry | This study | N/A |
| D. melanogaster: UASz-Pav-Dendra2 | This study | N/A |
| D. melanogaster: Jupiter-GFP | Buszczak et al.52 | CB05190 |
| D. melanogaster: SfGFP-Tum | Bloomington Drosophila stock center | RRID:BDSC_76264 |
| D. melanogaster: SfGFP-Sti | Bloomington Drosophila stock center | RRID:BDSC_81646 |
| D. melanogaster: Sqh-GFP | Bloomington Drosophila stock center | RRID:BDSC_57145 |
| D. melanogaster: mRFP-Anillin | Bloomington Drosophila stock center | RRID:BDSC_52220 |
| D. melanogaster: Shrub-GFP | Bloomington Drosophila stock center | RRID:BDSC_32559 |
| D. melanogaster: UASp-GFP-Alix | Eikenes et al.44 | N/A |
| D. melanogaster: 20X UAS-mC3PA-GFP | Pfeiffer et al.51 | N/A |
| Mouse: CD-1 | Kaelyn Sumigray, Yale | N/A |
| Hydra vulgaris: Kiel AEP | Celina Juliano, UC Davis | N/A |
| Oligonucleotides | ||
| Pav Forward: CCTCCGCTTCCACCGGAACCTCCTTTCGACTTCTTGCTGCT | This study | N/A |
| Pav Reverse: TGCCCAGCCAGGTGTGGTAACGAGGGTACCACTAGTTCTA | This study | N/A |
| Dendra2 Forward: CCGGTGGAAGCGGAGGTAGCGGCATGAACACCCCGGGAATTAACC | This study | N/A |
| Dendra2 Reverse: TAGAACTAGTGGTACCCTCGTTACCACACCTGGCTGGGCA | This study | N/A |
| Recombinant DNA | ||
| UASz vector | DeLuca and Spradling54 | RRID:DGRC_1431 |
| UASz-pav-dendra2 | This study | N/A |
| Software and Algorithms | ||
| Fiji | Schindelin et al.59 | RRID:CVCL_0299 |
| Prism | https://www.graphpad.com/scientific-software/prism/ | RRID:SCR_002798 |
| Qtiplot | www.qtiplot.com | N/A |
| Adobe Illustrator | https://www.adobe.com/uk/products/illustrator.html | RRID:SCR_010279 |
| Other | ||
| 35mm glass bottom dishes | MatTek | P35G-1.5-1.4-C |
Highlights.
Midbody core remodeling precedes formation of transient and stable germline ring canals
Ring canals originate from stabilized midbody rings
Aberrant midbody remodeling impacts ring canal formation and protein movement
Abscission inhibition stabilizes germ cell ring canals after they form
Acknowledgements
We thank Pier Paolo D’Avino for the anti-Sticky antibody and Kaisa Haglund and Damien Brünner for critical reagents. We thank Celina Juliano for providing Hydra vulgaris animals and technical advice. We are also grateful to Kaelyn Sumigray for providing mice, technical advice, and access to the Leica Stellaris laser scanning confocal microscope. We sincerely thank Chia-Ling Hsieh for performing mouse testis dissections. Lastly, we would like to thank members of the Cooley and Sumigray labs for helpful discussions as well as Andrew Cox for critical reading of the manuscript. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank the DRSC/TRiP Functional Genomics Resources at Harvard Medical School for providing transgenic RNAi fly stocks used in this study. The Hts/Adducin 1B1 (generated by H.D. Lipschitz) and α-Tubulin 4A1 (generated by M.T. Fuller) monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by the National Institutes of Health (F32GM136029 to K.L.P, and R01GM043301 and R35GM141961 to L.C.). K.L.P also received support from the Surdna Foundation and Yale Venture Fund.
Footnotes
Competing Interests
The authors declare no competing interests.
Inclusion and Diversity
We support inclusive, diverse and equitable conduct of research.
Supplemental Information
Supplemental Information includes seven figures and four movies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- 1.Haglund K, Nezis IP, and Stenmark H (2011). Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 4, 1–9. 10.4161/cib.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu K, Jensen L, Lei L, and Yamashita YM (2017). Stay Connected: A Germ Cell Strategy. Trends Genet. 33, 971–978. 10.1016/j.tig.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthel D, and Detmer A (1990). The spermatogenesis of Halichondria panicea (Porifera, Demospongiae). Zoomorphology 110, 9–15. 10.1007/BF01632807. [DOI] [Google Scholar]
- 4.Fawcett DW, Ito S, and Slautterback D (1959). The occurrence of intercellular bridges in groups of cells exhibiting. J. Biophys. Biochem. Cytol. 5, 453–460. 10.1083/jcb.5.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dym M, and Fawcett DW (1971). Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol. Reprod. 4, 195–215. 10.1093/biolreprod/4.2.195. [DOI] [PubMed] [Google Scholar]
- 6.Greenbaum MP, Iwamori T, Buchold GM, and Matzuk MM (2011). Germ cell intercellular bridges. Cold Spring Harb. Perspect. Biol. 3, 1–18. 10.1101/cshperspect.a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertho S, Clapp M, Banisch TU, Bandemer J, Raz E, and Marlow FL (2021). Zebrafish dazl regulates cystogenesis and germline stem cell specification during the primordial germ cell to germline stem cell transition. Dev. 148. 10.1242/dev.187773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D, and Cooley L (1996). Stable intercellular bridges in development: the cytoskeleton lining the tunnel. Trends Cell Biol. 6, 474–479. [DOI] [PubMed] [Google Scholar]
- 9.Mullins JM, and Biesele JJ (1977). Terminal phase of cytokinesis in D-98S cells. J. Cell Biol. 73, 672–684. 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Avino PP (2017). Citron kinase - renaissance of a neglected mitotic kinase. J. Cell Sci. 130, 1701–1708. 10.1242/jcs.200253. [DOI] [PubMed] [Google Scholar]
- 11.Agromayor M, and Martin-Serrano J (2013). Knowing when to cut and run: Mechanisms that control cytokinetic abscission. Trends Cell Biol. 23, 433–441. 10.1016/j.tcb.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Fededa JP, and Gerlich DW (2012). Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14, 440–447. 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- 13.Green RA, Paluch E, and Oegema K (2012). Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58. 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 14.Mahowald AP (1971). The formation of ring canals by cell furrows in Drosophila. Zeitschrift für Zellforsch. und Mikroskopische Anat. 118, 162–167. 10.1007/BF00341561. [DOI] [PubMed] [Google Scholar]
- 15.Koch EA, and King RC (1969). Further studies on the ring canal system of the ovarian cystocytes of Drosophila melanogaster. Zeitschrift für Zellforsch. und Mikroskopische Anat. 102, 129–152. 10.1007/BF00336421. [DOI] [PubMed] [Google Scholar]
- 16.Iwamori T, Iwamori N, Ma L, Edson MA, Greenbaum MP, and Matzuk MM (2010). TEX14 Interacts with CEP55 To Block Cell Abscission. Mol. Cell. Biol. 30, 2280–2292. 10.1128/mcb.01392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenbaum MP, Ma L, and Matzuk MM (2007). Conversion of midbodies into germ cell intercellular bridges. Dev. Biol. 305, 389–396. 10.1016/j.ydbio.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hime GR, Brill JA, and Fuller MT (1996). Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 109, 2779–2788. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita YM (2018). Subcellular specialization and organelle behavior in germ cells. Genetics 208, 19–51. 10.1534/genetics.117.300184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong SK, Foote C, and Tan C (2010). Mutations of DMYPT cause over constriction of contractile rings and ring canals during Drosophila germline cyst formation. Dev. Biol. 346, 161–169. 10.1016/j.ydbio.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Adams RR, Tavares AAM, Salzberg A, Bellen HJ, and Glover DM (1998). pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12, 1483–1494. 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman RS, Price KL, Mannix KM, Ayers KM, Hudson AM, and Cooley L (2020). Drosophila sperm development and intercellular cytoplasm sharing through ring canals do not require an intact fusome. Dev. 147. 10.1242/dev.190140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu CK, Coughlin M, and Mitchison TJ (2012). Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell 23, 1024–1034. 10.1091/mbc.E11-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elia N, Sougrat R, Spurlin TA, Hurley JH, and Lippincott-Schwartz J (2011). Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U. S. A. 108, 4846–4851. 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy RW, Tokuyasu KT, Lindsley DL, and Garavito M (1979). The Germinal Proliferation Center in the Testis of Drosophila melanogaster. J. Ultrasructure Res. 69, 180–190. 10.7868/s0869565218110257. [DOI] [PubMed] [Google Scholar]
- 26.Gönczy P, and Dinardo S (1996). The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122, 2437–2447. 10.1242/dev.122.8.2437. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Tiyaboonchai A, Yamashita YM, and Hunt AJ (2011). Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development 138, 831–837. 10.1242/dev.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng X, and Matunis E (2011). Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138, 3367–3376. 10.1242/dev.065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong SK, and Tan C (2010). Germline cyst formation and incomplete cytokinesis during Drosophila melanogaster oogenesis. Dev. Biol. 337, 84–98. 10.1016/j.ydbio.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Kuznetsov S, Lyanguzowa M, and Bosch TCG (2001). Role of epithelial cells and programmed ceil death in Hydra spermatogenesis. Zoology 104, 25–31. 10.1078/0944-2006-00005. [DOI] [PubMed] [Google Scholar]
- 31.Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, and Juliano CE (2019). Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science (80-.). 365. 10.1126/science.aav9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kechad A, Jananji S, Ruella Y, and Hickson GRX (2012). Anillin acts as a bifunctional linker coordinating midbody ring biogenesis during cytokinesis. Curr. Biol. 22, 197–203. 10.1016/j.cub.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Amine N, Carim SC, Wernike D, and Hickson GRX (2019). Rho-dependent control of the Citron kinase, Sticky, drives midbody ring maturation. Mol. Biol. Cell 30, 2185–2204. 10.1091/mbc.E19-04-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Amine N, Kechad A, Jananji S, and Hickson GRX (2013). Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J. Cell Biol. 203, 487–504. 10.1083/jcb.201305053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenhart KF, and DiNardo S (2015). Somatic Cell Encystment Promotes Abscission in Germline Stem Cells following a Regulated Block in Cytokinesis. Dev. Cell 34, 192–205. 10.1016/j.devcel.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Avino PP, and Capalbo L (2016). Regulation of midbody formation and function by mitotic kinases. Semin. Cell Dev. Biol. 53, 57–63. 10.1016/j.semcdb.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Bassi ZI, Audusseau M, Riparbelli MG, Callaini G, and D’Avino PP (2013). Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc. Natl. Acad. Sci. U. S. A. 110, 9782–9787. 10.1073/pnas.1301328110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naim V, Imarisio S, Di Cunto F, Gatti M, and Bonaccorsi S (2004). Drosophila citron kinase is required for the final steps of cytokinesis. Mol. Biol. Cell 15, 5053–5063. 10.1091/mbc.E04-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Cunto F, Imarisio S, Camera P, Boitani C, Altruda F, and Silengo L (2002). Essential role of citron kinase in cytokinesis of spermatogenic precursors. J. Cell Sci. 115, 4819–4826. 10.1242/jcs.00163. [DOI] [PubMed] [Google Scholar]
- 40.Eda M, Yonemura S, Kato T, Watanabe N, Ishizaki T, Madaule P, and Narumiya S (2001). Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J. Cell Sci. 114, 3273–3284. 10.1242/jcs.114.18.3273. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe S, De Zan T, Ishizaki T, and Narumiya S (2013). Citron kinase mediates transition from constriction to abscission through its coiled-coil domain. J. Cell Sci. 126, 1773–1784. 10.1242/jcs.116608. [DOI] [PubMed] [Google Scholar]
- 42.Lie-Jensen A, Ivanauskiene K, Malerød L, Jain A, Tan KW, Laerdahl JK, Liestøl K, Stenmark H, and Haglund K (2019). Centralspindlin Recruits ALIX to the Midbody during Cytokinetic Abscission in Drosophila via a Mechanism Analogous to Virus Budding. Curr. Biol. 29, 3538–3548.e7. 10.1016/j.cub.2019.09.025. [DOI] [PubMed] [Google Scholar]
- 43.Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, and Hurley JH (2008). Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278–1286. 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eikenes ÅH, Malerød L, Christensen AL, Steen CB, Mathieu J, Nezis IP, Liestøl K, Huynh JR, Stenmark H, and Haglund K (2015). ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division In Vivo. PLoS Genet. 11. 10.1371/journal.pgen.1004904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathieu J, Michel-Hissier P, Boucherit V, and Huynh JR (2022). The deubiquitinase USP8 targets ESCRT-III to promote incomplete cell division. Science (80-.). 376, 818–823. 10.1126/science.abg2653. [DOI] [PubMed] [Google Scholar]
- 46.Robinson DN, Cant K, and Cooley L (1994). Morphogenesis of Drosophila ovarian ring canals. Development 120, 2015–2025. 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- 47.Echard A, Hickson GRX, Foley E, and O’farrell PH (2004). Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14, 1685–1693. 10.1016/j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, and King N (2011). Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 357, 73–82. 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoops H, Nishii I, Kirk D, Baluska F, Volkmann D, and Barlow P (2006). Cytoplasmic Bridges in Volvox and Its Relatives. In Cell-Cell Channels, pp. 65–84. 10.1007/978-0-387-46957-7_4. [DOI] [Google Scholar]
- 50.Chaigne A, and Brunet T (2022). Incomplete abscission and cytoplasmic bridges in the evolution of eukaryotic multicellularity. Curr. Biol. 32, R385–R397. 10.1016/j.cub.2022.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Jankovics F, and Brunner D (2006). Transiently Reorganized Microtubules Are Essential for Zippering during Dorsal Closure in Drosophila melanogaster. Dev. Cell 11, 375–385. 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 52.Pfeiffer BD, Truman JW, and Rubin GM (2012). Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 109, 6626–6631. 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. (2007). The Carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531. 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karpova N, Bobinnec Y, Fouix S, Huitorel P, and Debec A (2006). Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton 63, 301–312. 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- 55.Deluca SZ, and Spradling AC (2018). Efficient expression of genes in the Drosophila germline using a UAS promoter free of interference by Hsp70 piRNAs. Genetics 209, 381–387. 10.1534/genetics.118.300874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris LX, and Spradling AC (2011). Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development 138, 2207–2215. 10.1242/dev.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Avino PP, Savoian MS, and Glover DM (2004). Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J. Cell Biol. 166, 61–71. 10.1083/jcb.200402157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lenhoff HM, and Brown RD (1970). Mass culture of Hydra: an improved method and its application to other aquatic invertebrates. Lab. Anim. 4, 139–154. 10.1258/002367770781036463. [DOI] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ring canal formation in the Drosophila testis occurs via a midbody-like intermediate. Related to Figure 1.
A time-lapse movie of a testis expressing Pav::mCherry (expressed at endogenous levels; magenta) and His::GFP (green). A dividing two-cell cyst is outlined in a white box. The testis is oriented to the left (apical tip) and the hub is evidenced by the accumulation of Pav::mCherry foci. The movie is displayed at a frame rate of 5 frames per second.
Midbody formation, but not reorganization, in dividing cyst stem cells in the Drosophila testis. Related to Figure 1.
A time-lapse movie of a testis expressing Pav::mCherry (expressed at endogenous levels; magenta) and His::GFP (green). A dividing cyst stem cell is marked with a white arrow. The hub is evidenced by the accumulation of Pav::mCherry foci. The movie is displayed at a frame rate of 5 frames per second.
Ring canal formation in the germline stem cell occurs via a midbody-like intermediate. Related to Figure 1.
A time-lapse movie of a testis expressing Pav::GFP (expressed at endogenous levels; green) and mRFP::Anillin (driven by nos-Gal4, magenta). A dividing germline stem cell is marked with a white arrow. The testis is oriented to the left (apical tip) and the hub is evidenced by the accumulation of mRFP::Anillin foci. The movie is displayed at a frame rate of 5 frames per second. The movie captures the dynamics of Pav and Anilin from the formation of a cleavage furrow to the beginning of collapse of the mature midbody.
Ring canal formation in the Drosophila ovary occurs via a midbody-like intermediate. Related to Figure 3.
A time-lapse movie of a germarium expressing Pav::mCherry (expressed at endogenous levels; magenta) and His::GFP (green). The anterior end of the germarium is oriented down and left. Top panel: merge of Pav::GFP (magenta) and His::RFP (green). Bottom panel: individual channel of Pav::GFP. The frame rate is 5 frames per second.
Data Availability Statement
Plasmids, fly lines, and microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Raw data from Figure S5 was deposited on Mendeley at https://data.mendeley.com/datasets/4bdxw3v44s/draft?a=49439b09-22c3-4da4-a02f-596c371c514a
Any additional information required to reanalyze the data reported is available from the lead contact upon request.


