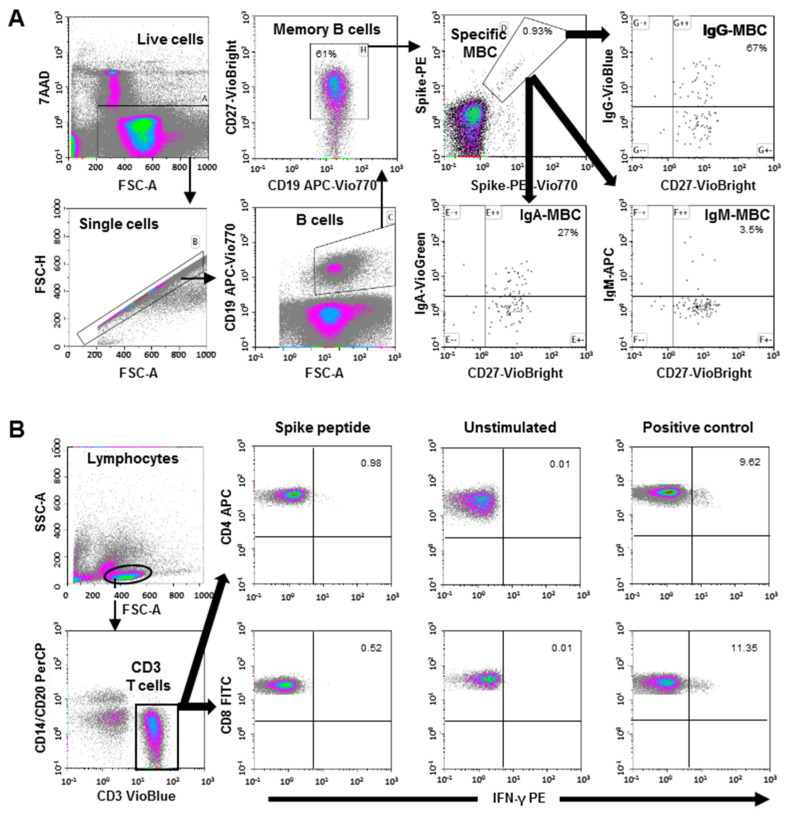
Figure 1.
(A) Flow cytometry strategy for the quantification of SARS-CoV-2-specific memory B-cells (MBC). Viable cells (FSC-A/7AAA plot) were plotted with FSC-H/FSC-A parameters to exclude doublets. Single cells were then gated using CD19-APC-Vio770 and CD27-Vio-Bright-FITC to identify memory B-cells. Spike-specific B-cells were then identified with a double staining with the two spike-tetramer on the diagonal of the dot plot. Finally, the use of IgG-VioBlue, IgA-VioGreen, and IgM-APC were used to quantify each specific isotype of spike-specific memory B-cells. The results are recorded as percentage among the total memory B-cells and isotypes among specific memory B-cells. (B) Flow cytometry gating strategy for the quantification of SARS-CoV-2 spike-specific T-cells. After stimulation with the spike peptide, staining of the cells was carried out with the following fluorochrome-conjugated antibodies: CD3-VioBlue, CD4-APC, CD8-FITC, CD14-PerCP, CD20-PerCP, IFN-γ-PE, and FcR blocking reagent. To exclude dead cells, viability 405/520 fixable dye staining was added for the last 10 min of incubation. The samples were measured and analyzed by flow cytometry on a MACSQuant Analyzer 10 using MACSQuantify software. At least 105 cells were analyzed and gated with the following strategy: Single (FSC-A/FSC-H dot plot) and live cells were first selected. Cell debris, monocytes, and B-cells were excluded from the analysis with CD14-and CD20-PerCP antibodies. Then, lymphocytes were selected with an FSC-A/SSC-A dot plot, and CD3 T-cells were gated. IFN-γ expression was finally analyzed separately for CD4+ and CD8+ T-cells. A representative sample of negative control (without stimulation), positive control (stimulated with SEB), and with SARS-CoV-2 spike peptide are shown.