Abstract
Targeting critical epigenetic regulators reverses aberrant transcription in cancer, thereby restoring normal tissue function1–3. The interaction of menin with lysine methyltransferase 2A (KMT2A), an epigenetic regulator, is a dependence in acute leukaemia caused by either rearrangement of KMT2A or mutation of the nucleophosmin 1 gene (NPM1)4–6. KMT2A rearrangements occur in up to 10% of acute leukaemias and have an adverse prognosis, whereas NPM1 mutations occur in up to 30%, forming the most common genetic alteration in acute myeloid leukaemia7,8. Here, we describe the results of the first-in-human phase 1 clinical trial investigating revumenib (SNDX-5613), a potent and selective oral inhibitor of the menin–KMT2A interaction, in patients with relapsed or refractory acute leukaemia (ClinicalTrials.gov, NCT04065399). We show that therapy with revumenib was associated with a low frequency of grade 3 or higher treatment-related adverse events and a 30% rate of complete remission or complete remission with partial haematologic recovery (CR/CRh) in the efficacy analysis population. Asymptomatic prolongation of the QT interval on electrocardiography was identified as the only dose-limiting toxicity. Remissions occurred in leukaemias refractory to multiple previous lines of therapy. We demonstrate clearance of residual disease using sensitive clinical assays and identify hallmarks of differentiation into normal haematopoietic cells, including differentiation syndrome. These data establish menin inhibition as a therapeutic strategy for susceptible acute leukaemia subtypes.
Subject terms: Acute myeloid leukaemia, Targeted therapies
Revumenib, a potent and selective oral inhibitor of the menin–KMT2A interaction, is associated with a low frequency of treatment-related adverse events and promising clinical activity in patients with relapsed or refractory acute leukaemia.
Main
The prognosis of acute leukaemias harbouring rearrangements of the gene lysine methyltransferase 2A (KMT2A), previously known as mixed-lineage leukaemia (MLL), is poor, with a 5-year overall survival of less than 25% (ref. 7). KMT2A rearrangements (KMT2Ar) occur in 80% of infant acute lymphoblastic leukaemia (ALL) and in 5–15% of children and adults with acute leukaemia, whether myeloid, lymphoid or mixed phenotype9. Mutated nucleophosmin 1 gene (NPM1) is the most common genetic alteration in adult acute myeloid leukaemia (AML), occurring in up to 30% of patients8. Currently there are no targeted therapies specifically approved for acute leukaemia with KMT2Ar or mutated NPM1.
Genetic rearrangements of KMT2A lead to aberrant expression of homeobox (HOX) genes and their DNA-binding cofactor MEIS1 (ref. 10). This gene expression programme, normally expressed in stem cells, causes a haematopoietic differentiation block and leukaemic transformation11. For leukaemias driven by KMT2Ar, menin is a critical oncogenic cofactor4. The menin-binding motif is preserved throughout all KMT2A fusion proteins, and menin is an essential cofactor for binding to HOX gene promoters7. Similarly, in AML with mutated NPM1, the interaction between wild-type KMT2A and menin leads to HOX- and MEIS1-mediated leukaemogenic transcription6,12. Blockade of the menin–KMT2A interaction disrupts the assembly of oncogenic KMT2A wild-type or fusion complexes on chromatin12. Preclinical studies showed that menin inhibition downregulates HOX and MEIS1 transcription and reverses leukaemogenesis in KMT2Ar-r- or NPM1-mutated leukaemia models5,12,13. Revumenib, previously known as SNDX-5613, is a potent, oral, selective inhibitor of the menin–KMT2A interaction. Treatment with revumenib led to abrogation of aberrant HOX gene expression and dramatic antileukaemic activity in those models (Extended Data Fig. 1; GEO accession no. for RNA sequencing (RNA-seq) data, GSE216730). In this first-in-human clinical trial we assessed the safety, maximum tolerated dose (MTD), recommended phase 2 dose (RP2D) and pharmacokinetic and pharmacodynamic profiles in patients with relapsed or refractory acute leukaemia, and present the clinical activity of revumenib in patients with KMT2Ar or mutated NPM1.
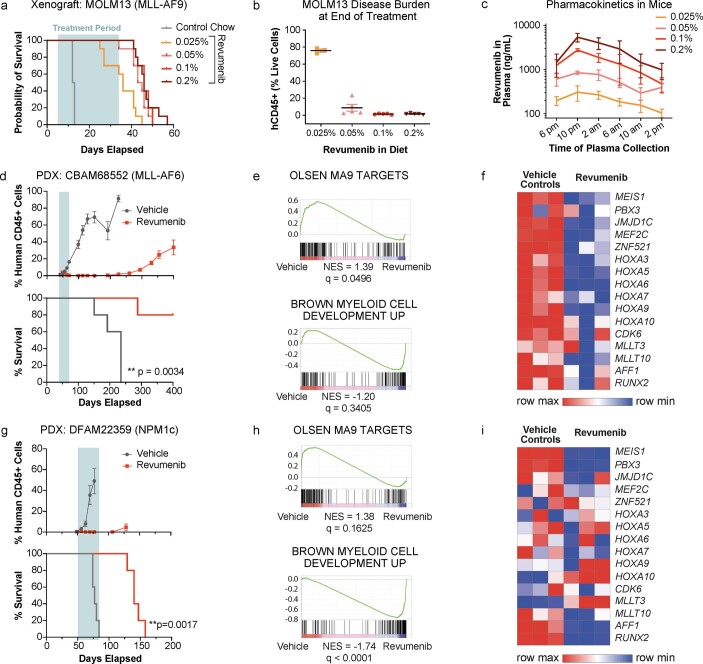
Extended Data Fig. 1. Revumenib suppresses both KMT2Ar and NPM1-mutant AML in preclinical models of leukaemia.
NSG mice were engrafted with the KMT2Ar cell line MOLM-13 (a; n = 10) or patient derived xenografts (PDX) harbouring either a KMT2Ar (d; n = 5) or NPM1 mutation (g; n = 5). Mice were treated for 28 days with revumenib, which was formulated in chow, across a dose range of 0.025% to 0.2% (a) or at 0.1% fixed dose (d, g). Leukaemic burden (CD45+) was assessed at end of treatment (b; 0.025%: n = 3; 0.05-0.2%: n = 5, data represent mean ± SEM) or throughout the study (d, g; data represent mean ± SEM). For MOLM-13 engrafted mice, revumenib showed clear dose-dependent exposure (c; n = 3 with 3 individual measurements per timepoint and dose, data represent mean ± SD), which translated into dose-responsive effect on survival benefit (a) and leukaemic burden at end of treatment (b). Similarly, revumenib treatment of the PDX models led to significant suppression of leukaemic burden and significant survival benefit in each (d, g). P-values were determined by log-rank (Mantel–Cox) tests. Adjustments were not made for multiple comparisons. Revumenib treatment also led to broad changes in the transcriptional program (f, i; n = 3 per treatment group; GEO accession number for RNAseq data, GSE216730) with GSEA results consistent with previously reported signatures (e, h; n = 3 per treatment group). GSEA, gene set enrichment analysis; NES, normalized enrichment score; NSG, NOD scid gamma; PDX, patient-derived xenograft; SD, standard deviation; SEM, standard error of the mean.
This phase 1 dose-escalation study was conducted across nine sites in the United States. Because revumenib is a substrate of cytochrome P450 3A4 (CYP3A4), two parallel dose-escalation cohorts, one without (Arm A) and one with (Arm B) strong CYP3A4 inhibitors, were conducted. Revumenib was administered orally every 12 h (q12h) in continuous 28-day cycles. Although all patients could enrol at study initiation regardless of cytogenetic and mutational profile, given the strong preclinical rationale demonstrating that the menin–KMT2A interaction is a targetable vulnerability in acute leukaemia with KMT2Ar or mutated NPM1, an early amendment to the protocol restricted enrolment to patients with relapsed or refractory acute leukaemia with KMT2Ar or mutated NPM1 (88% of the total study population). Between 5 November 2019 and 31 March 2022, a total of 68 patients (37 in Arm A and 31 in Arm B) were enroled and treated. The data cutoff date for this analysis was 31 March 2022. Patient disposition is presented in Extended Data Fig. 2.
Extended Data Fig. 2. CONSORT diagram and patient disposition on trial.
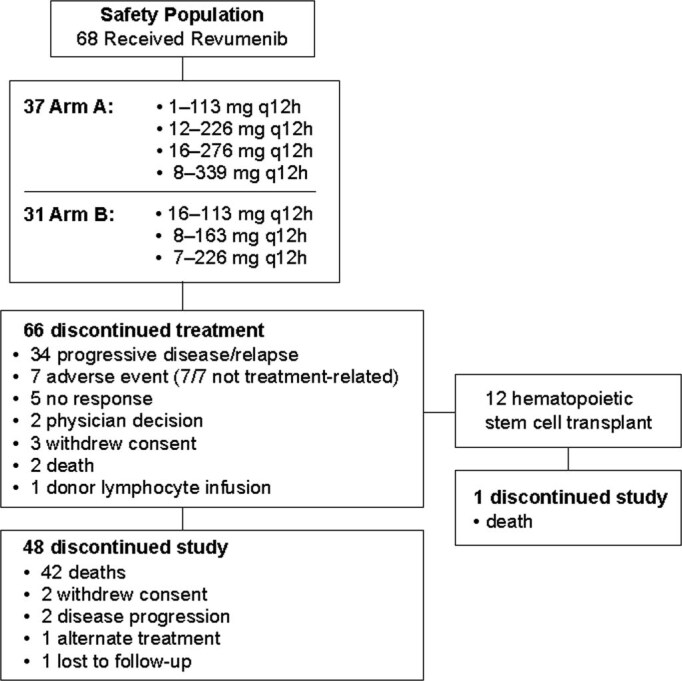
The CONSORT diagram shows the number of patients from Arms A and B who discontinued treatment and lists the reasons for treatment and study discontinuation.
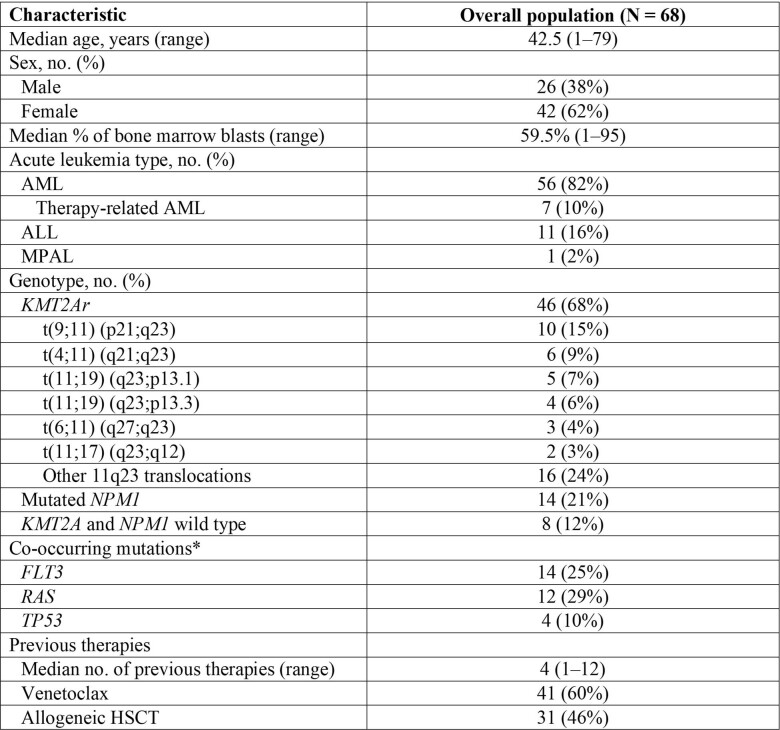
Baseline characteristics of patients are provided in Extended Data Table 1. There were 56 patients (82%) with relapsed or refractory AML, 11 (16%) with ALL and one with mixed-phenotype acute leukaemia (2%). Forty-six patients (68%) had KMT2Ar, 14 (21%) had mutated NPM1 and eight (12%) had neither KMT2Ar nor NPM1 mutations. Sixty patients were adults (at least 18 years old) and eight were children or adolescents (under 18 years of age). The median age was 42.5 years (range, 0.8–79). The median age of adult patients was 50.5 years (range, 19–79) and the median age among paediatric patients was 2.5 years (range, 0.8–16). The study population was heavily pretreated with a median of four previous lines of therapy (range, 1–12), and 31 patients (46%) had relapsed after an allogenic stem cell transplant. All patients who received at least one dose of revumenib were included in the safety analysis whereas only patients with KMT2Ar or mutated NPM1 were included in the efficacy analysis.
Extended Data Table 1.
Baseline characteristics
*In patients for whom co-occurring mutation data were available and assessed at each site.
AML, acute myeloid leukaemia; ALL, acute lymphoblastic leukaemia; FLT3, fms-related tyrosine kinase; HSCT, haematopoietic stem cell transplant; MPAL, mixed phenotype acute leukaemia; RAS, rat sarcoma virus.
All values are shown as n (%) except otherwise indicated.
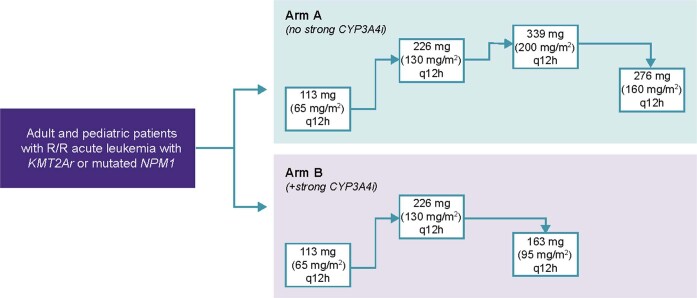
The dose-escalation schema is shown in Extended Data Fig. 3. The only dose-limiting toxicity observed in both Arm A and Arm B was grade 3 prolongation of the QT interval (over 500 ms) on electrocardiography (ECG), a toxicity predicted in preclinical animal studies. These events occurred without any clinical correlate or symptoms. This dose-limiting toxicity was observed in Arm A at dose levels of 226 mg q12h (n = 1) and 339 mg q12h (n = 2), and in Arm B at 113 mg q12h (n = 1) and 226 mg q12h (n = 2). The incidence of grade 3 QT prolongation was 10% at dose levels either at or below MTD.
Extended Data Fig. 3. Dose escalation schema.
The revumenib dose was adjusted based on the body surface area (BSA) for patients weighing less than 40 kg as indicated for each corresponding dose level shown in parentheses. CYP3A4i, cytochrome P450 3A4 inhibitor; q12h, every 12 h; R/R, relapsed or refractory.
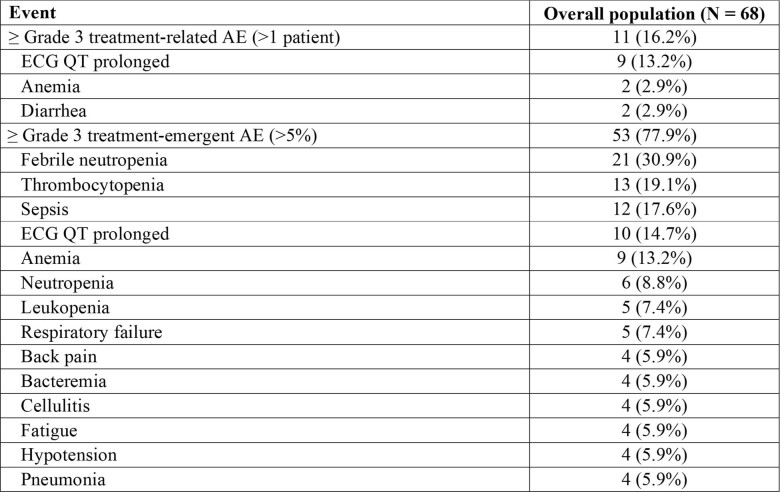
A total of 67 treated patients (99%) had an adverse event during treatment with revumenib whereas 53 patients (78%) had a treatment-related adverse event (TRAE) of any grade (Table 1). The most frequent treatment-emergent adverse events (TEAEs) of any grade, irrespective of a relationship to revumenib, were prolongation of the QT interval (56%), nausea (50%), vomiting (40%) and febrile neutropenia (31%); a full listing of these adverse events in at least 20% of the patients is given in Table 1. The most frequent TEAEs of grade 3 or higher were febrile neutropenia (31%), thrombocytopenia (19%) and sepsis (18%) (Extended Data Table 2). The most frequent TRAE was prolongation of the QT interval, in 36 patients (53%). Eleven patients (16%) had grade 3 or 4 TRAEs (Extended Data Table 2). The most frequent grade 3 TRAE was prolongation of the QT interval, in nine patients (13%). There were no ventricular arrythmias, and all episodes of grade 3 QT prolongation resolved either before the next dose or with protocol-directed dose hold. All patients with grade 3 QT prolongation were able to resume dosing at the next-lower dose level. Other grade 3 TRAEs included diarrhoea (3%), hypercalcaemia (2%), tumour lysis syndrome (2%), anaemia (3%), asthenia (2%), fatigue (2%), neutropenia (2%), thrombocytopenia (2%), and hypokalaemia (2%). Grade 4 TRAEs included neutropenia (2%) and thrombocytopenia (2%), with one event each. No patients permanently discontinued revumenib due to TRAEs, and no TRAEs led to death.
Table 1.
Any-grade treatment-related and TEAEs, regardless of causality
| Event | Overall population (n = 68) |
|---|---|
| Any-grade TRAE (5% or over) | 53 (77.9%) |
| ECG QT prolonged | 36 (52.9%) |
| Nausea | 18 (26.5%) |
| Differentiation syndrome | 11 (16.2%) |
| Vomiting | 11 (16.2%) |
| Diarrhoea | 7 (10.3%) |
| Decreased appetite | 5 (7.4%) |
| Dysgeusia | 5 (7.4%) |
| Any-grade TEAE (20% or over) | 67 (98.5%) |
| ECG QT prolonged | 38 (55.9%) |
| Nausea | 34 (50.0%) |
| Vomiting | 27 (39.7%) |
| Febrile neutropenia | 21 (30.9%) |
| Diarrhoea | 20 (29.4%) |
| Fatigue | 18 (26.5%) |
| ALT increased | 17 (25.0%) |
| Headache | 16 (23.5%) |
| Hyperphosphataemia | 16 (23.5%) |
| Hypokalaemia | 15 (22.1%) |
| Hyponatraemia | 15 (22.1%) |
| Thrombocytopenia | 15 (22.1%) |
| Epistaxis | 14 (20.6%) |
| Peripheral oedema | 14 (20.6%) |
All AEs shown as n (%).
ALT, alanine aminotransferase.
Extended Data Table 2.
Grade 3 or higher treatment-related and treatment-emergent adverse events
AE, adverse event; ECG, electrocardiogram.
Adverse events are shown as n (%).
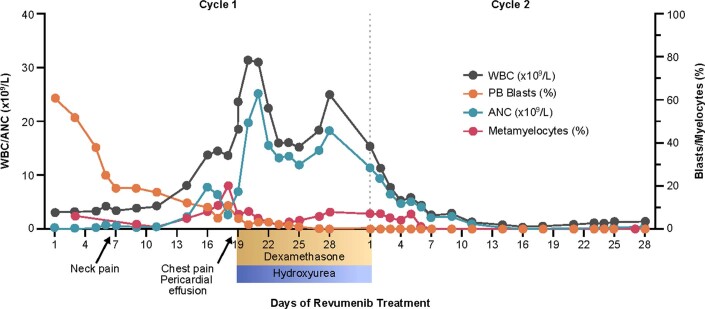
A common occurrence in antileukaemia therapies that induce myeloid differentiation is the development of a clinically apparent differentiation syndrome. Differentiation syndrome is caused by cytokine alterations associated with haematopoietic differentiation, with common manifestations including fever, arthralgias, leukocytosis, pleural or pericardial effusions and respiratory or renal failure in severe cases14,15. This had been mostly reported in patients with myeloid leukaemias treated with all-trans retinoic acid, arsenic trioxide and isocitrate dehydrogenase inhibitors14–16. In this study, differentiation syndrome was reported in 11 patients (16%), all grade 2. All cases of differentiation syndrome resolved following treatment with corticosteroids, with the addition of hydroxyurea for associated leukocytosis seen in five of the 11 patients (Extended Data Figs. 4 and 5). One patient missed one day of dosing, but otherwise treatment with revumenib was uninterrupted because of differentiation syndrome. The median time of onset of differentiation syndrome was 18 days (range, 5–41) with manifestations including bone pain or arthralgia, pericardial effusion, pleural effusion, pulmonary infiltrates, increase in creatinine, rash, oedema and pyrexia.
Extended Data Fig. 4. Morphologic evidence of myeloid differentiation.
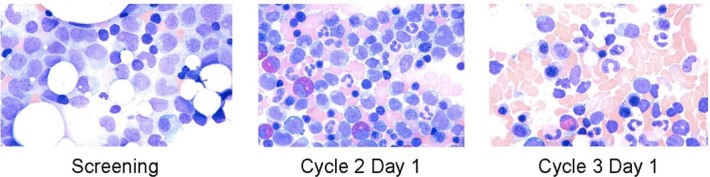
Photomicrographs of bone marrow biopsies demonstrating morphologic evidence of myeloid differentiation in a patient who achieved complete remission; magnification is 40x.
Extended Data Fig. 5. Changes in peripheral blood during differentiation syndrome are associated with the menin inhibitor revumenib.
Example from a 71-year-old patient with KMT2Ar AML relapsed after an allogeneic stem cell transplant, who received revumenib at 339 mg PO q12h (Arm A), and achieved CRh, MRD negative remission. Differentiation syndrome manifested as chest pain with a small pericardial effusion, and a possible prodrome of neck pain likely related to expansion of cervical nodes, all resolved promptly with initiation of steroids followed by tapering doses. Hydroxyurea was used to control leukocytosis. AML, acute myeloid leukaemia; ANC, absolute neutrophil count; CRh, complete remission with partial haematologic recovery; MRD, minimal or measurable residual disease; PB, peripheral blood; PO, by mouth; q12h, every 12 h; WBC, white blood cell.
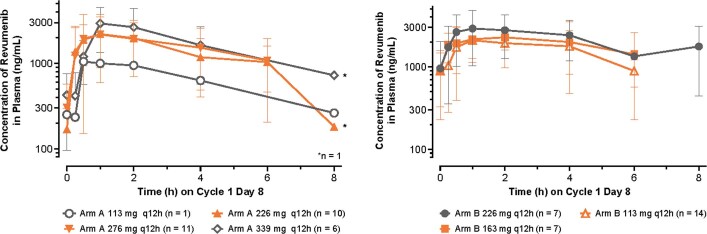
Pharmacokinetic studies showed that dose-proportional exposure was achieved in both Arm A (no strong CYP3A4 inhibitor) and Arm B (with a strong CYP3A4 inhibitor). Steady-state levels were achieved in approximately 48 h, with no evidence of drug accumulation (Extended Data Fig. 6).
Extended Data Fig. 6. Dose proportional exposure was achieved across both arms.
The half-life of revumenib in Arm A (without strong CYP3A4 inhibitors) was approximately 3 h at the cycle 1 day 8 assessment of the 276-mg q12h dose level and was approximately 8 h at the same assessment in Arm B (with a strong CYP3A4 inhibitor) at the 163-mg q12h dose level. Data represent mean ± SD. Data cutoff date for pharmacokinetic analysis was July 11, 2022. CYP3A4, cytochrome P450 3A4; q12h, every 12 h; SD, standard deviation.
Following a review of prespecified protocol criteria on safety, tolerability and pharmacokinetic data, the revumenib doses of 226 mg q12h and 276 mg q12h in Arm A and 113 mg q12h and 163 mg q12h in Arm B met the prespecified criteria for RP2D.
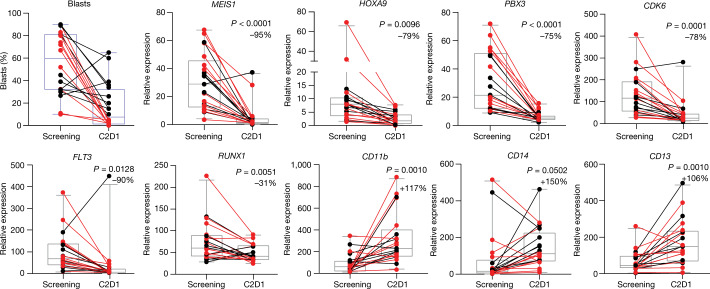
The pharmacodynamic effects of revumenib were assessed through transcriptional changes following treatment using RNA-seq of bone marrow cells (Fig. 1). Consistent with the established mechanism of action, menin inhibition using revumenib resulted in downregulation of the critical leukaemogenic target genes MEIS1, homeobox A9 (HOXA9), pre-B-cell leukaemia transcription factor 3 (PBX3) and cyclin-dependent kinase 6 (CDK6), and an increase in expression of genes associated with differentiation, such as integrin alpha M (CD11b) and CD14. We also observed transcriptional suppression of fms-like tyrosine kinase 3 (FLT3) following treatment, a putative transcriptional target of MEIS1; FLT3 is frequently mutated in multiple subtypes of AML, including KMT2Ar AML (10%), and is particularly common in AML with mutated NPM1 (60%)8,17–19.
Fig. 1. Transcriptional changes following treatment with the menin inhibitor revumenib in patients with relapsed or refractory acute leukaemia with KMT2Ar or mutated NPM1.
RNA-seq before and after treatment with revumenib, showing downregulation of critical leukaemogenic target genes MEIS1, HOXA9 and PBX3 and increase in expression of genes associated with differentiation (CD11b, CD14), with transcriptional suppression of FLT3, a putative transcriptional target of MEIS1. The change in bone marrow blast percentage following treatment is shown. Box plots represent median gene expression or median bone marrow blast percentage, and the 95% CI along with percentage change in gene expression following treatment. Responders are shown in red, nonresponders in black. Results were obtained using a paired t-test with a two-sided P value. Adjustments were not made for multiple comparisons. This analysis included a cohort of 21 evaluable patients. Revumenib was administered in continious 28-day cycles. C2D1, day 1 of treatment cycle 2.
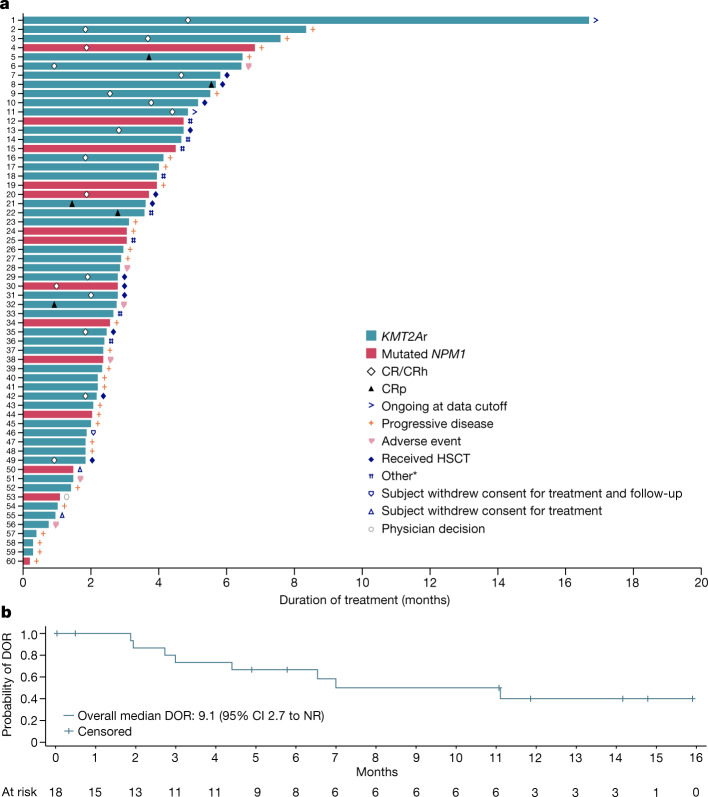
The rate of complete remission or complete remission with partial haematologic recovery (CR/CRh) was 30% (95% confidence interval (CI) 18.8–43.2) in 18 of 60 evaluable patients with undetectable measurable residual disease (MRD), as assessed by multiparameter flow cytometry, in 14 of 18 patients (78%) who achieved CR/CRh (Fig. 2a and Table 2). The overall response rate (CR/CRh/complete remission with incomplete platelet recovery (CRp)/complete remission with incomplete haematologic recovery (CRi)/morphologic leukaemia-free state) was 53% (32 of 60 evaluable patients). The median time to CR/CRh was 1.9 months (range, 0.9–4.9). Although imaging assessment was not mandated on study, responses were notably seen in both bone marrow and extramedullary sites in two of six evaluable patients with extramedullary leukaemia at enrolment, with representative imaging shown in Extended Data Fig. 7. In an exploratory descriptive analysis, we evaluated responses by both leukaemia lineage and the adult and paediatric (under 18 years of age) populations. Morphologic remissions were identified in 27 of 49 patients (55%) with AML (95% CI 40.2–69.3), in four of ten patients (40%) with ALL (95% CI 12.2–73.8) and in the one patient with a mixed-phenotype acute leukaemia. Morphologic remission was noted in four of eight paediatric patients (50%; 95% CI 15.7–84.3) and in 28 of 52 adults (54%; 95% CI 39.5–67.8).
Fig. 2. Characterization of remissions with the menin inhibitor revumenib in susceptible relapsed or refractory acute leukaemia subtypes.
a, Time to response, duration of treatment (censored at time of HSCT) and patient status by the cutoff date. *Other reasons for treatment discontinuation included no response, relapse, death and donor lymphocyte infusion. b, Kaplan–Meier curve of duration of response (DOR) in patients with CR or CR/CRh without censoring at the time of an allogeneic stem cell transplant performed in 12 of 18 evaluable patients.
Table 2.
Responses to treatment
| Response | Efficacy population (n = 60) | KMT2Ar (n = 46) | Mutated NPM1 (n = 14) |
|---|---|---|---|
| Overall response* | 32 (53%) | 27 (59%) | 5 (36%) |
| Median time to first morphologic response (range), months | 0.95 (0.9–3.7) | 0.95 (0.9–3.7) | 0.99 (1.0–1.9) |
| Best response* | |||
| CR/CRh | 18 (30%) | 15 (33%) | 3 (21%) |
| CR | 12 (20%) | 9 (20%) | 3 (21%) |
| CRh | 6 (10%) | 6 (13%) | 0 |
| Median time to CR or CRh (range), months | 1.9 (0.9–4.9) | 2.0 (0.9–4.9) | 1.9 (1.0–1.9) |
| CRi | 0 | 0 | 0 |
| CRp | 5 (8%) | 5 (11%) | 0 |
| MLFS | 9 (15%) | 7 (15%) | 2 (14%) |
| Partial remission | 0 | 0 | 0 |
| No response | 19 (32%) | 12 (26%) | 7 (50) |
| Progressive disease | 7 (12%) | 6 (13%) | 1 (7%) |
| Missing | 2 (3%) | 1 (2%) | 1 (7%) |
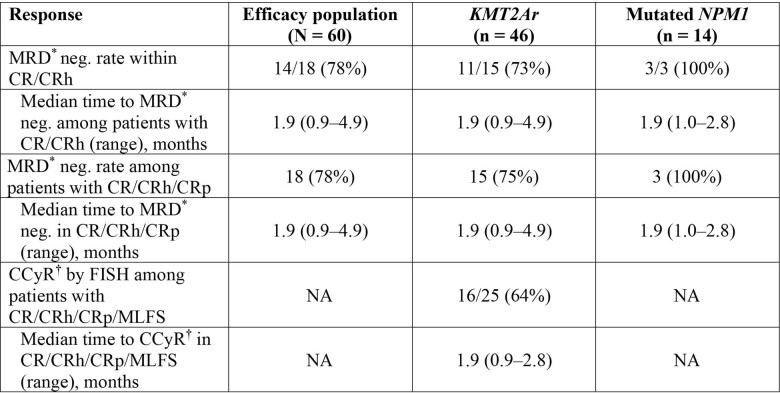
| MRD† neg. rate within CR/CRh | 14/18 (78%) | 11/15 (73%) | 3/3 (100%) |
| Median time to MRD† neg. among patients with CR/CRh (range), months | 1.9 (0.9–4.9) | 1.9 (0.9–4.9) | 1.9 (1.0–2.8) |
*Responses were assessed by the investigators; responses and MRD-negative rates are shown as n (%).
†MRD, minimal or measurable residual disease assessed at participating sites by either multicolour flow cytometry or PCR; MRD status percentage based on patients with non-missing MRD status out of all responders.
MLFS, morphologic leukaemia-free state.
Extended Data Fig. 7. Response in extra-medullary disease.
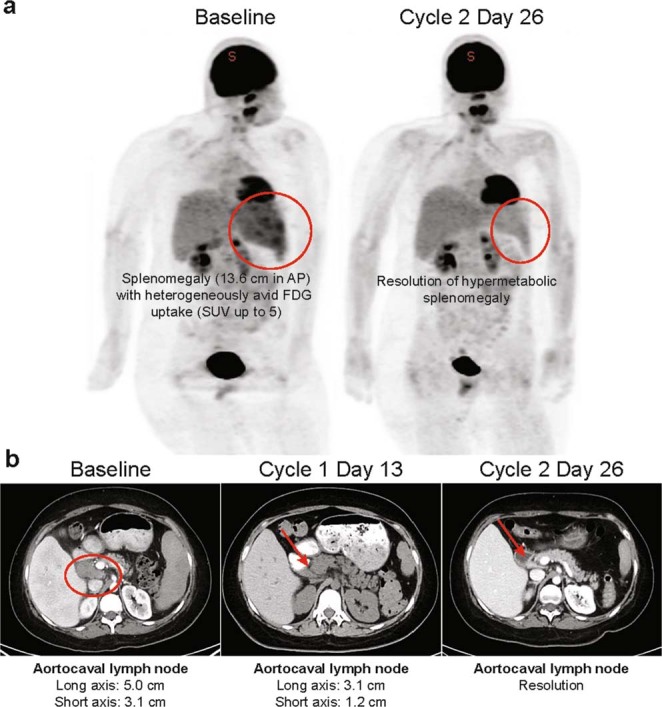
A. PET scan at baseline and after 2 cycles of treatment. B. Computed tomographic scans of target lesions and spleen from a 19-year-old with relapsed KMT2Ar AML, previously treated with three prior lines of therapy including 2 allogeneic stem cell transplants and local radiation to the spleen and abdominal nodes, received revumenib at 276 mg PO q12h (Arm A), achieved CRh, MRD negative remission with resolution of extramedullary disease in abdominal nodes and spleen. AML, acute myeloid leukaemia; AP, anterior-posterior; CRh, complete remission with partial haematologic recovery; D, day; FDG, fluorodeoxyglucose; MRD, minimal or measurable residual disease; PET, positron emission tomography; PO, by mouth; q12h, every 12 h; SUV, standardized uptake value.
In patients with KMT2Ar who achieved morphologic remission following the first cycle of therapy with revumenib, 21 of 25 (84%) retained the detectable fusions causing KMT2Ar during concomitant cytogenetic analyses. However, most patients later had clearance of these KMT2A rearrangements with subsequent cycles of therapy. The rate of complete cytogenetic response in patients with KMT2Ar who achieved morphologic clearance of their myeloblasts was 16 of 25 (64%) (Extended Data Table 3); these typically occurred after morphologic remission and, in some cases, after MRD negativity as assessed by immunophenotyping via flow cytometry. The median time to achieving complete cytogenetic response was 1.9 months (range, 0.9–2.8). These findings are consistent with the mechanism of action of differentiation agents, for which response leads first to a phenotypic differentiation of cells such that they retain the genetic fusion but are without aberrant morphology or leukaemia surface marker expression (as detected by flow cytometry)20,21. Subsequently with additional cycles following phenotypic remission, fusions are no longer detected by fluorescence in situ hybridization, probably because of apoptosis of differentiated cells or replacement by normal haematopoiesis.
Extended Data Table 3.
Measurable residual disease and cytogenetic responses
*MRD, minimal or measurable residual disease assessed at participating sites by multicolor flow cytometry or by polymerase chain reaction. MRD status percentage is based on patients with non-missing MRD status out of the responders. MRD-negative rates are shown as n (%).
†CCyR denotes complete cytogenetic remission in which KMT2A fusions were not detectable by fluorescence in situ hybridization in evaluable patients assessed at participating sites.
CR, complete remission; CRh, complete remission with partial haematologic recovery; CRi, complete remission with incomplete haematologic recovery; CRp, complete remission with incomplete platelet recovery; FISH, fluorescence in situ hybridization; MLFS, morphologic leukaemia free state; MRD, minimal or measurable residual disease; NA, not applicable given absence of disease defining cytogenetic abnormalities.
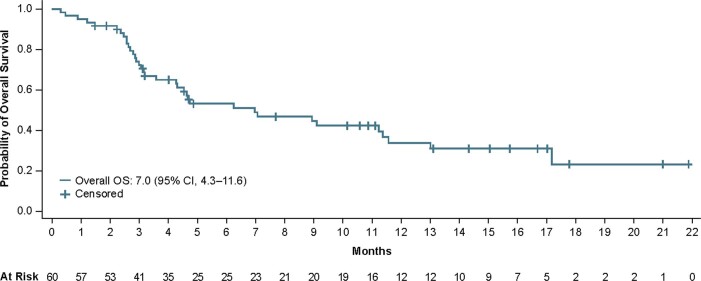
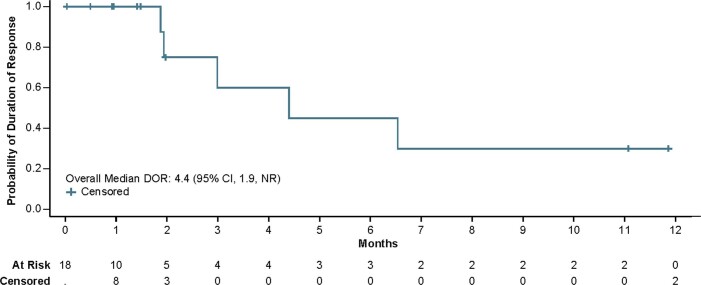
With a median follow-up of 11.9 months (95% CI 4.9–14.8) in patients who achieved CR/CRh, the median duration of response was 9.1 months (95% CI 2.7 to not reached (NR)) (Fig. 2b). Median overall survival in the efficacy population, regardless of remission status, was 7 months (95% CI 4.3–11.6) (Extended Data Fig. 8) and median follow-up of overall survival in the efficacy population was 14.3 months (95% CI 10.6–16.7). Twelve patients received an allogeneic stem cell transplant as consolidation following response to revumenib, with nine in remission at the time of the data cutoff, seven of whom have been in remission for over 6 months. The median duration of response in those who achieved CR/CRh with censoring at the time of allogeneic stem cell transplant was 4.4 months (95% CI 1.9–NR) (Extended Data Fig. 9).
Extended Data Fig. 8. Overall survival in patients with KMT2Ar or mutated NPM1.
OS, overall survival.
Extended Data Fig. 9. Kaplan-Meier curve of duration of response in patients with complete remission or complete remission with partial haematologic recovery with censoring at time of the allogeneic stem cell transplant.
DOR, duration of response; NR, not reached.
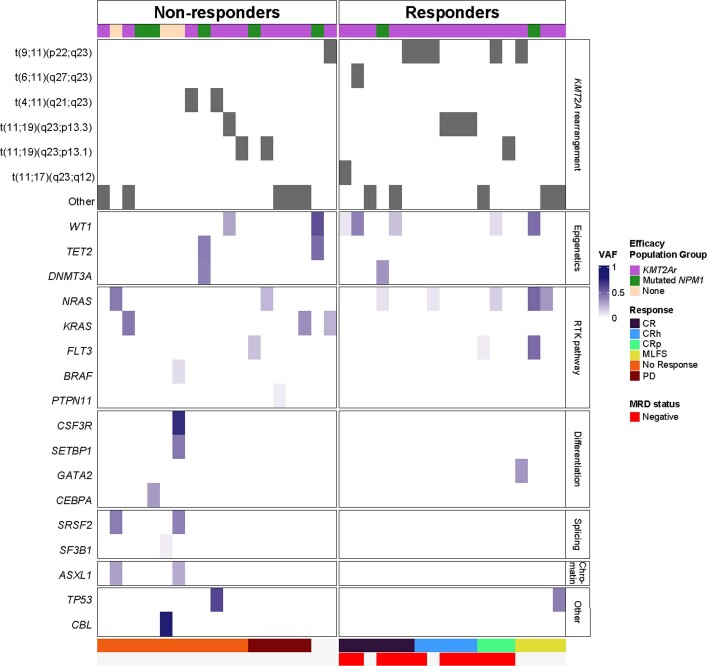
Consistent with the preclinical hypothesis regarding the efficacy of menin inhibition in patients with NPM1 mutations or KMT2Ar, there were no responses in the eight patients enroled on study who lacked either a KMT2Ar or NPM1 mutation. In evaluable patients for whom a targeted sequencing panel was performed at baseline, no specific KMT2A translocation or comutation was clearly associated with response in a limited analysis (Extended Data Fig. 10). Notably among evaluable patients with KMT2Ar or mutated NPM1, 14 had a comutation in FLT3, of whom 13 had been treated with a FLT3 inhibitor before enrolment. Among the 11 patients with comutated NPM1 and FLT3, three responded whereas two of the three patients with KMT2Ar and concomitant FLT3 mutations achieved a response. All responders had previously received a FLT3 inhibitor.
Extended Data Fig. 10. Mutation analysis and response.
This cohort included 37 patients evaluable for this analysis. CR, complete remission; CRh, complete remission with partial haematologic recovery; CRp, complete remission with incomplete platelet recovery; MLFS, morphologic leukaemia-free state; NR, not reached; PD, progressive dVAF, variant allele frequency.
Discussion
The treatment of patients who have relapsed or refractory acute leukaemia with KMT2Ar is challenging. The rate of CR or complete remission with incomplete count recovery in relapsed KMT2Ar AML following two previous lines of therapy is 9% (ref. 18). Despite notable progress in the treatment of childhood acute leukaemia, infant KMT2Ar acute leukaemias have remained a therapeutic challenge with high rates of resistance to multi-agent chemotherapy22,23. Mutations in NPM1 are the most common alterations in AML, and targeting of vulnerabilities associated with this subset expand the reach of targeted therapy to the largest portion of patients with this disease. The menin inhibitor revumenib has the potential to address these unmet needs. In this first-in-human clinical trial, we provide clinical evidence of the effectiveness of menin inhibition with an oral targeted therapy, which is the first epigenetic therapy that evicts protein complexes from chromatin, leading to remissions in patients with acute leukaemia. We found an encouraging clinical benefit, with deep molecular remissions and minimal toxicities, in a heavily pretreated population of both children and adults with advanced acute leukaemia. However, recent data suggest that clinical resistance to targeted menin inhibition is mediated through the acquisition of mutations in menin that prevent inhibitor binding and can lead to clinical relapse24. The emergence of resistance mutations highlights the critical role of the menin–KMT2A interaction in the pathogenesis of KMT2Ar and NPM1-mutant acute leukaemia.
Because revumenib disrupts the effect of epigenetic regulators in leukaemia that are dependent on the menin–KMT2A interaction, this leads to abrogation of aberrant gene expression and removal of the haematopoietic differentiation block12,13. In this study, we show evidence of this mechanism of action with differentiation of leukaemia cells, persistence of cytogenetic abnormalities at the time of morphologic response and differentiation syndrome associated with menin inhibition in some patients, which resolved following appropriate therapy.
Other leukaemia genotypes have recently been identified as susceptible to menin inhibition, such as AML with rearrangement of the nucleoporin 98 gene (NUP98), a common and adverse genotype among children with relapsed and refractory disease25. Another example is the rare occurrence of AML with translocations involving the meningioma-1 gene (MN1)26. Therefore, menin inhibitors have the potential to reach a larger subset of acute leukaemia with similar dependency on the menin–KMT2A interaction, which could be identified using precision approaches testing characteristic gene expression.
In conclusion, in children and adults with highly refractory acute leukaemia with KMT2Ar or NPM1 mutation, menin inhibition with revumenib monotherapy was associated with promising antileukaemic activity leading to deep and sustained remission.
Methods
Study design and oversights
We conducted a phase 1, multicentre, open-label, dose-escalation study (ClinicalTrials.gov, NCT04065399). Revumenib was administered orally in either capsule or liquid formulation, q12h, in 28-day continuous cycles with dose adjustment by body surface area for patients weighing under 40 kg (Extended Data Fig. 3). We employed a rolling-6 design, an algorithm-based extension of the 3+3 design that allows for concurrent accrual of two to six patients onto a dose level, thereby shortening the duration of phase 1 without increasing the risk of toxicity27. Therefore, the number of patients enrolled followed this dose-escalation design without an absolute prior estimation of sample size. The study included two parallel dose-escalation cohorts, for patients either not taking (Arm A) or taking (Arm B) strong CYP3A4 inhibitors. Dose expansion occurred at safe and efficacious doses. Dose-limiting toxic effects were defined as nonhaematologic toxic effects of grade 3 or higher during cycle 1, or as haematologic toxicities directly attributed to the study drug rather than to the underlying disease.
The study was conducted in accordance with the principles of the Declaration of Helsinki and the International Council for Harmonization Good Clinical Practice. The protocol and amendments were approved by the institutional review board or ethics committee at The University of Texas MD Anderson Cancer Center, City of Hope, Washington University School of Medicine in St. Louis, Dana-Farber Cancer Institute, Winship Cancer Institute, Emory University School of Medicine, University of Chicago, Florida Cancer Specialists/Sarah Cannon Research Institute, University of Iowa and Memorial Sloan Kettering Cancer Center. All patients provided written informed consent. This study was designed by the sponsor (Syndax Pharmaceuticals) in collaboration with the investigators. The data were collected by the investigators and their research staff and were analysed by the sponsor and authors. Drafts of the manuscript were written by the first and last authors and revised in collaboration with all authors and the sponsor, all of whom vouch for the completeness and accuracy of the data and analyses, and for the adherence of the study to the protocol. Assistance in manuscript preparation for submission was provided by a professional service and paid for by the sponsor.
Patients
An early amendment to the protocol allowed patients aged 30 days and older to be enroled on this study; additionally, it restricted the eligibility criteria from any relapsed or refractory acute leukaemia to only those with KMT2Ar or mutated NPM1 in Arms A and B of the study. Mutational status was assessed at each site.
Study assessments
The primary objectives of the phase 1 portion of this study were to assess safety, the MTD or, if different, RP2D and to characterize the pharmacokinetic parameters of revumenib in arms based on concomitant CYP3A inhibitors. Exploratory objectives included assessment of antileukaemic activity in the efficacy population, which consisted of patients with KMT2Ar or mutated NPM1. All study assessments and analyses included both paediatric and adult patients.
Adverse events were graded with use of the Common Terminology Criteria for Adverse Events, v.5.0. Clinical efficacy was assessed by the investigators with use of a modified version of the 2017 European LeukemiaNet response criteria, to additionally include CRh28. Guidelines for managing prolongation of the QT interval included electrolyte repletion and adjustment of revumenib dose. Investigators were encouraged to administer corticosteroids following suspicion of differentiation syndrome based on guidelines included in the protocol. Measurable residual disease assessment was performed in participating institutions using multicolour flow cytometry or PCR29–31. In an exploratory mutational analysis we used the ArcherDx VariantPlex Myeloid Panel, a 75-gene, next-generation targeted sequencing product examining genes frequently mutated in myeloid and lymphoid malignancies. Powered by Anchored Multiple PCR chemistry, the panel enables deep strand-specific amplification of unique molecular barcoded DNA fragments for sequencing. Archer Analysis software was used for the analysis and interpretation of sequencing data in the detection of single-nucleotide variants and indels. Variant calling was performed using Invitae’s Comprehensive Targeted Mutation File (v.1.6), and the Vision variant caller to report well-classified variants.
Statistical analysis
All patients who received at least one dose of revumenib were included in the safety analysis, whereas only those with KMT2Ar or mutated NPM1 were included in the efficacy analysis. Time-to-event end points were estimated using the Kaplan–Meier method. Descriptive statistics were used for other clinical, laboratory and pharmacokinetic variables. Clinical data were captured in Medidata Classic Rave 2021.2.0, and data analyses were performed using SAS v.9.4 and GraphPad Prism v.8.0. Changes in gene expression were analysed using a paired t-test.
RP2D was determined by the safety review committee based on review of pharmacokinetics, safety and tolerability data among evaluable patients.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-023-05812-3.
Supplementary information
Acknowledgements
This study was funded by Syndax Pharmaceuticals. We thank the patients, their families and caregivers for participating in the study, in addition to all of the participating investigators and research staff. Bioinformatics support was provided by Fios Genomics. G.C.I. was additionally supported by the K12 Paul Calabresi Clinical Scholarship Award (no. NIH/NCI K12 CA088084) and E.M.S. by NIH/NCI Cancer Center Support Grant no. P30 CA008748 to Memorial Sloan Kettering Cancer Center.
Extended data figures and tables
Author contributions
G.C.I., E.M.S., G.R., M.L. M. and Y.G. conceptualized and designed the study. G.C.I. and E.M.S. prepared the first draft of the manuscript, provided critical review and revisions of the drafts and approved the decision to submit for publication. F.P., S.A.A. and J.A.P. designed, performed and analysed preclinical experiments using patient-derived xenografts. G.C.I., I.A., J.D.P., B.C., R.S., M.A., M.J.T., M.R.P., D.S.D., S.Shenoy, N.S., H.K. and E.M.S. enroled and treated patients. G.C.I., I.A., J.D.P., B.C., R.S., M.A., M.J.T., M.R.P., D.S.D., S.Shenoy, N.S., S.A.A., F.P., J.A.P., G.R., R.G.B., M.L.M., P.O., Y.G., V.K., S.Smith, G.M.M. and E.M.S. collected data, analysed and interpreted the results and reviewed and edited manuscript drafts. S.Smith participated in conception and design analysis and interpretation of results. M.R.P. and S.A.A. participated in trial conduct and clinical trial discussions. G.M.M. conceptualized and designed the study, analysed data and interpreted results for preclinical studies. V.K. collected data and analysed and interpreted results for pharmacodynamic studies. All authors have read and agreed on the content of the manuscript.
Peer review
Peer review information
Nature thanks Jorge Cortes, Eunice Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Syndax Pharmaceuticals (Syndax) is committed to providing qualified scientific researchers access to anonymized data and clinical study reports from the company’s clinical trials for the purpose of conducting legitimate scientific research. Syndax is also obligated to protect the rights and privacy of trial participants and, as such, has a procedure in place for evaluating and fulfilling requests for sharing company clinical trial data with qualified external scientific researchers. Following submission of a request (to datarequest@syndax.com), Syndax will provide an outline of the process and requirements for submitting a data request. Feasible requests will be reviewed by a committee of Syndax subject matter experts to assess the scientific validity of the request and the qualifications of the requestors. In line with data privacy legislation, submitters of approved requests must enter into a standard data-sharing agreement with Syndax before Syndax may grant data access. Data will be made available for request after product approval in the United States and European Union, or after product development is discontinued. There are circumstances that may prevent Syndax from sharing requested data, including country- or region-specific regulations. If Syndax declines the request, it will communicate the decision to the investigator. Access to genetic or exploratory biomarker data requires a detailed statistical analysis plan that is collaboratively developed by the requester and Syndax subject matter experts.
Competing interests
G.C.I. received consultancy or advisory role fees from Novartis, Kura Oncology and NuProbe and received research funding from Celgene, Novartis, Kura Oncology, Syndax Pharmaceuticals, Merck, Cullinan Oncology and NuProbe. I.A. received consultancy or advisory role fees from Amgen, Pfizer, Jazz, AbbVie and Agios, research funding from AbbVie and Macrogenics and honoraria from Amgen, Pfizer, Jazz, AbbVie and Agios. J.D.P. has a consultancy role with Incyte and RiverVest Venture Partners, has served as a board member or advisory committee member for RiverVest Venture Partners, Magenta, hC Bioscience, Inc. and WUGEN, has received research funding from NeoImmune Tech, Macrogenics, Incyte, Bioline Rx and WUGEN and holds patents or pending patents for UCART7 for treatment of T-ALL, VLA-4 inhibitors for stem cell mobilization and NT-I7 for CART expansion. R.S. has served on the steering committee of AbbVie and advisory boards of AbbVie, AvenCell, CTI Pharma, Kura One, Genentech, Actinium, Arog, BMS, Boston Pharmaceuticals, GSK, Janssen, Jazz, Novartis, Syros, Takeda, Elevate Bio, Syndax Pharmaceuticals, Gemoab, BerGenBio, Foghorn Tera, Aprea, Innate, Actinium and OncoNova; served as DSMB for Aptevo, Epizyme, Takeda and Syntrix/ACI Clinical; on the focus group of BerGenBio; and on AML Expert Council of GSK and Grand Rounds of Jazz Pharmaceuticals. M.A. has served on the advisory boards of Kite Pharma and Syndax Pharmaceuticals. M.J.T. has a consulting or advisory role with AbbVie and CVS, has an expert testimony role with Apotex and received research funding from AbbVie, Gilead Sciences, Janssen, Merck, Pharmacyclics, Syndax Pharmaceuticals, TG Therapeutics and Tolero. M.R.P. served in a leadership role with ION Pharma; received honoraria from Pfizer, Pharmacyclics, Bayer, Janssen Oncology, Genentech and Adaptive Biotechnologies; has a consulting or advisory role with Pharmacyclics/Janssen and Pfizer/EMD Serono; served on the Speakers’ Bureau of Exelixis, Genentech/Roche, Taiho Pharmaceutical and Celgene; and received research funding from Acerta Pharma, ADC Therapeutics, Agenus, Aileron Therapeutics, AstraZeneca, BioNTech AG, Boehringer Ingelheim, Celgene, Checkpoint Therapeutics, CicloMed, Clovis Oncology, Cyteir Therapeutics, Daiichi Sankyo, Lilly, EMD Serono, Evelo Therapeutics, FORMA Therapeutics, Genentech/Roche, Gilead Sciences, GlaxoSmithKline, H3 Biomedicine, Hengrui Therapeutics, Hutchison MediPharma, Ignyta, Incyte, Jacobio, Janssen, Klus Pharma, Kymab, Loxo, LSK Biopartners, Lycera, Macrogenics, Merck, Millennium, Mirati Therapeutics, Moderna Therapeutics, Pfizer, Placon, Portola Pharmaceuticals, Prelude Therapeutics, Ribon Therapeutics, Seven and Eight Biopharmaceuticals, Syndax Pharmaceuticals, Taiho Pharmaceutical, Takeda, Tesaro, TopAlliance BioSciences, Inc., Vigeo, ORIC, Artios, IgM Biosciences, Puretech, BioTheryX, Black Diamond Therapeutics, IgM Biosciences, NGM Biopharmaceuticals, Nurix, PureTech, Relay Therapeutics, Samumed, Silicon Therapeutics, TeneoBio, Treadwell Therapeutics, Zymeworks, Olema, Adagene, Astellas, NGM, Accutar Biotech, TeneoBio, Novartis, Compugen, Black Diamond Therapeutics, MabSpace Biosciences, Immunogen and Blueprint Pharmaceuticals. D.S.D. has a consulting or advisory role with Tempus, Inc. S. Shenoy has a consulting or advisory role with Artio, BMS and Takaeda. H.K. received honoraria/advisory board/consulting fees from AbbVie, Amgen, Amphista, Ascentage, Astellas, Biologix, Curis, Ipsen Biopharmaceuticals, KAHR Medical, Labcorp, Novartis, Pfizer, Shenzhen Target Rx, Stemline and Takeda; and received research funding from AbbVie, Amgen, Ascentage, BMS, Daiichi Sankyo, Immunogen, Jazz, and Novartis. S.A.A. received stock or other ownership from Neomorph, Inc., C4 Therapeutics, Cyteir Therapeutics, Accent Therapeutics and Mana Therapeutics; has a consulting or advisory role with Neomorph, Inc., C4 Therapeutics, Cyteir Therapeutics, Accent Therapeutics, Mana Therapeutics and Twentyeight-Seven Therapeutics; received research funding from Syndax Pharmaceuticals and Janssen; and holds patents, royalties and other intellectual property for MENIN inhibition in NPM1 AML: WO/2017/132398A1. G.R. is a former employee of Syndax Pharmaceuticals and a current employee of Boston Pharmaceuticals. R.G.B. is an employee of Syndax Pharmaceuticals and has stock or other ownership at Syndax Pharmaceuticals. M.L.M., P.O. and G.M.M. are employees of Syndax Pharmaceuticals. M.L.M. has a consulting or advisory role at Nuvalent, holds patents, royalties and other intellectual property at Syndax Pharmaceuticals and Nuvalent and has stock or other ownership at Syndax Pharmaceuticals and Johnson & Johnson. P.O. has a consulting or advisory role at Patrys and Twentyeight-Seven Therapeutics, holds patents, royalties and other intellectual property at Syndax Pharmaceuticals and has stock or other ownership at Syndax Pharmaceuticals. G.M.M. has a consulting or advisory role at Syndax Pharmaceuticals, holds patents, royalties and other intellectual property at Syndax Pharmaceuticals and has stock or other ownership at Syndax Pharmaceuticals. Y.G. is an employee of Syndax Pharmaceuticals and has stock or other ownership at Syndax Pharmaceuticals and AstraZeneca. S. Smith has a consultancy or advisory role with Syndax Pharmaceuticals. E.M.S. has a consulting or advisory role with Gilead, CTI Biopharma, Epizyme, AbbVie, Pinotbio, Neoleukin Genesis, Genentech, Jazz, Novartis, Celgene, Calithera, Takeda, Janssen, BMS, Kronos, Kura, Auron, Syndax Pharmaceuticals, Servier, Agios and Remix and received research funding from Biotheryx, Agios, Servier, Eisai, BMS, Bayer, Syndax, Syros and Loxo. B.C., F.P., J.A.P., N.S. and V.K. declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ghayas C. Issa, Email: gcissa@mdanderson.org
Eytan M. Stein, Email: steine@mskcc.org
Extended data
is available for this paper at 10.1038/s41586-023-05812-3.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-023-05812-3.
References
- 1.Lo-Coco F, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013;369:111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- 2.DiNardo CD, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 3.Stein EM, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–731. doi: 10.1182/blood-2017-04-779405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama A, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 5.Grembecka J, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kühn MW, et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov. 2016;6:1166–1181. doi: 10.1158/2159-8290.CD-16-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issa GC, et al. Therapeutic implications of menin inhibition in acute leukemias. Leukemia. 2021;35:2482–2495. doi: 10.1038/s41375-021-01309-y. [DOI] [PubMed] [Google Scholar]
- 10.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu BD, Hess JL, Horning SE, Brown GAJ, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 12.Uckelmann HJ, et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science. 2020;367:586–590. doi: 10.1126/science.aax5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krivtsov AV, et al. A menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell. 2019;36:660–673. doi: 10.1016/j.ccell.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanz MA, Montesinos P. How we prevent and treat differentiation syndrome in patients with acute promyelocytic leukemia. Blood. 2014;123:2777–2782. doi: 10.1182/blood-2013-10-512640. [DOI] [PubMed] [Google Scholar]
- 15.Fathi AT, et al. Differentiation syndrome associated with enasidenib, a selective inhibitor of mutant isocitrate dehydrogenase 2: analysis of a phase 1/2 study. JAMA Oncol. 2018;4:1106–1110. doi: 10.1001/jamaoncol.2017.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norsworthy KJ, et al. Differentiation syndrome with ivosidenib and enasidenib treatment in patients with relapsed or refractory IDH-mutated AML: a U.S. Food and Drug Administration systematic analysis. Clin. Cancer Res. 2020;26:4280–4288. doi: 10.1158/1078-0432.CCR-20-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dzama MM, et al. Synergistic targeting of FLT3 mutations in AML via combined menin-MLL and FLT3 inhibition. Blood. 2020;136:2442–2456. doi: 10.1182/blood.2020005037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issa GC, et al. Predictors of outcomes in adults with acute myeloid leukemia and KMT2A rearrangements. Blood Cancer J. 2021;11:162. doi: 10.1038/s41408-021-00557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bill M, et al. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proc. Natl Acad. Sci. USA. 2020;117:26340–26346. doi: 10.1073/pnas.2014732117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimwade D, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J. Clin. Oncol. 2009;27:3650–3658. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]
- 21.Andreeff M, et al. Hexamethylene bisacetamide in myelodysplastic syndrome and acute myelogenous leukemia: a phase II clinical trial with a differentiation-inducing agent. Blood. 1992;80:2604–2609. doi: 10.1182/blood.V80.10.2604.2604. [DOI] [PubMed] [Google Scholar]
- 22.Hilden JM, et al. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108:441–451. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pieters R, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370:240–250. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 24.Perner, F. et. al. MEN1 mutations mediate clinical resistance to menin inhibition. Nature10.1038/s41586-023-05755-9 (2023). [DOI] [PMC free article] [PubMed]
- 25.Heikamp EB, et al. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood. 2021;139:894–906. doi: 10.1182/blood.2021012806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libbrecht C, et al. Menin is necessary for long term maintenance of meningioma-1 driven leukemia. Leukemia. 2021;35:1405–1417. doi: 10.1038/s41375-021-01146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skolnik JM, et al. Shortening the timeline of pediatric phase I trials: the rolling six design. J. Clin. Oncol. 2008;26:190–195. doi: 10.1200/JCO.2007.12.7712. [DOI] [PubMed] [Google Scholar]
- 28.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buccisano F, et al. The kinetics of reduction of minimal residual disease impacts on duration of response and survival of patients with acute myeloid leukemia. Leukemia. 2006;20:1783–1789. doi: 10.1038/sj.leu.2404313. [DOI] [PubMed] [Google Scholar]
- 30.Terwijn M, et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J. Clin. Oncol. 2013;31:3889–3897. doi: 10.1200/JCO.2012.45.9628. [DOI] [PubMed] [Google Scholar]
- 31.Freeman SD, et al. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J. Clin. Oncol. 2013;31:4123–4131. doi: 10.1200/JCO.2013.49.1753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Syndax Pharmaceuticals (Syndax) is committed to providing qualified scientific researchers access to anonymized data and clinical study reports from the company’s clinical trials for the purpose of conducting legitimate scientific research. Syndax is also obligated to protect the rights and privacy of trial participants and, as such, has a procedure in place for evaluating and fulfilling requests for sharing company clinical trial data with qualified external scientific researchers. Following submission of a request (to datarequest@syndax.com), Syndax will provide an outline of the process and requirements for submitting a data request. Feasible requests will be reviewed by a committee of Syndax subject matter experts to assess the scientific validity of the request and the qualifications of the requestors. In line with data privacy legislation, submitters of approved requests must enter into a standard data-sharing agreement with Syndax before Syndax may grant data access. Data will be made available for request after product approval in the United States and European Union, or after product development is discontinued. There are circumstances that may prevent Syndax from sharing requested data, including country- or region-specific regulations. If Syndax declines the request, it will communicate the decision to the investigator. Access to genetic or exploratory biomarker data requires a detailed statistical analysis plan that is collaboratively developed by the requester and Syndax subject matter experts.