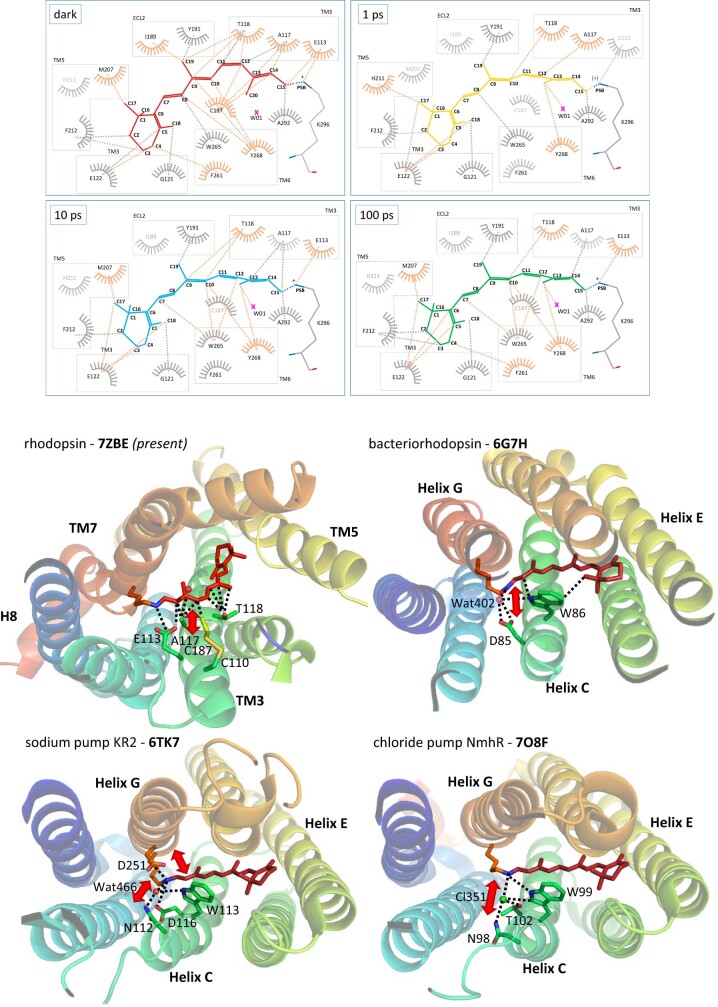
Extended Data Fig. 7. Interactions of retinal with its binding pocket at 0, 1, 10 and 100 ps of photoactivation and comparison with microbial rhodopsins.
LIGPLOT58 view of retinal interactions within the rhodopsin binding site (upper four panels) (set distance < 3.6 Å) at different time-delays of photoactivation, dark state, 1, 10 and 100 ps-photoactivated states. The amino acids and dashed lines of interatomic interactions labelled in orange are the site of major changes, showing new interactions, e.g. with water W01 or losing contact with C187(ECL2). Retinal binding pocket of non-homologous rhodopsins (lower four panels) (PDBid codes shown in bold) from mammalian (the rhodopsin GPCR from Bos taurus (this study) (PDBid=7ZBE)) and prokaryotes (bacteriorhodopsin proton pump from Halobacterium salinarum (PDBid=6G7H); KR2 sodium pump from Krokinobacter eikastus (PDBid=6TK7) and the NmHR chloride pump from Nonlabens marinus (PDBid=7O8F)). The third transmembrane helix (“TM3” in GPCRs and “helix C” in prokaryotes) has been described as a main interaction site for retinal104, often carrying the stabilizing counterion, like E113 in rhodopsin. Projected on the dark state structure of these four rhodopsins, some important structural rearrangements upon retinal isomerization (cis-to-trans for mammalian rhodopsin, trans-to-cis for prokaryotic rhodopsins), observed in TR-SFX studies in the picosecond range, are indicated with a red arrow. The disruption of these important interactions weakens the stabilization of the retinal chromophore by TM3/helix C.