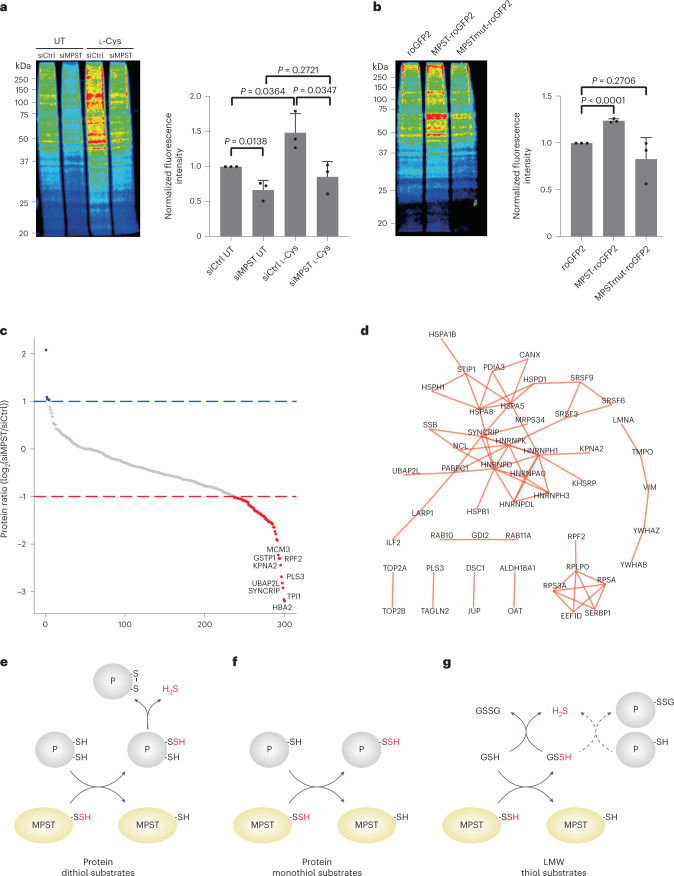
Fig. 6. MPST contributes to global protein persulfidation.
a, Overall persulfidation levels in HEK293 MSR cells before and after depletion of MPST (left panel). Cells were treated with 5 mM l-Cys for 30 min or were left untreated (UT). Relative persulfidation levels are indicated by Coomassie-normalized fluorescence intensity (right panel). Data are presented as mean and individual values (n = 3 biologically independent experiments) ± s.e.m. Statistical analysis based on a two-tailed unpaired t-test. b, Overall persulfidation levels in HEK293 MSR cells ectopically overexpressing roGFP2, MPST-roGFP2 or MPST(C248S)-roGFP2 (MPSTmut-roGFP2) (left panel). Relative persulfidation levels are indicated by Coomassie-normalized fluorescence intensity (right panel). Data are presented as mean and individual values (n = 3 biologically independent experiments) ± s.e.m. Statistical analysis based on a two-tailed unpaired t-test. c, Influence of MPST depletion on the persulfidation of individual proteins. Proteins depleted by at least twofold in MPST-depleted cells are marked in red. d, Interaction analysis of candidate MPST target proteins. Edges represent experimentally supported protein–protein interactions (confidence score >0.4) acquired from the STRING database50. The graph was generated with Cytoscape51. e–g, Summary of MPST-driven transpersulfidation. The MPST-bound persulfide (MPST-SSH) sulfurates thiol-containing molecules, the outcome depending on the type of acceptor. e, Sulfur transfer to proteins (P) with vicinal dithiols (roGFP2, Trx1) generates a protein disulfide and releases H2S. f, Sulfur transfer to protein monothiols leads to longer-lived protein persulfides. g, Sulfur transfer to GSH generates GSSH, which releases H2S to generate GSSG or (dotted lines) to glutathionylate proteins.