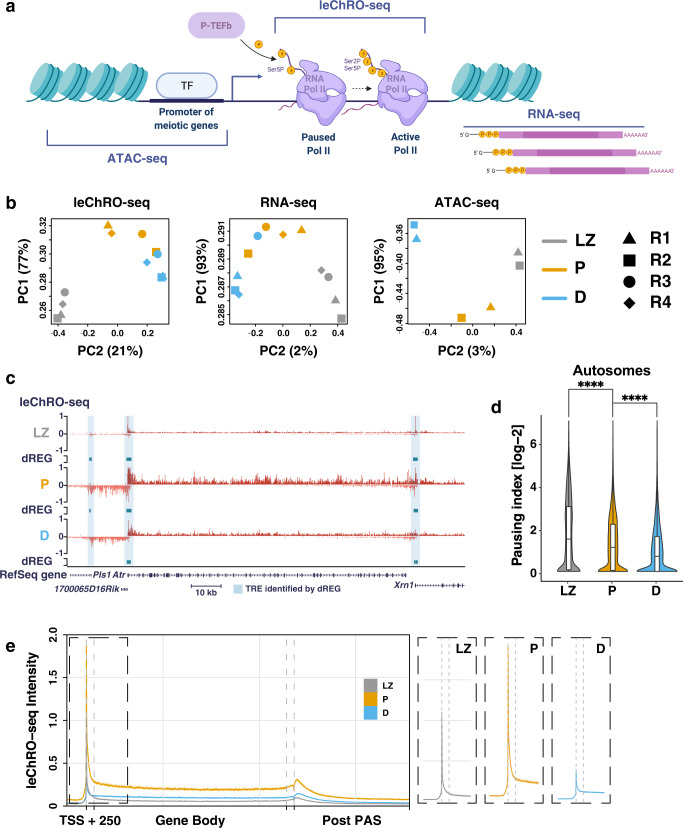
Fig. 2. leChRO-seq detects nascent transcription in prophase I spermatocytes.
a Schematic of key steps in the transcription cycle. The transcription start site (TSS) is labeled with an arrow; nucleosomes are depicted in teal; nascent RNA is shown in dark red; and mRNA is shaded in dark pink. ATAC-seq detects accessible regulatory elements, such as promoters and enhancers. leChRO-seq provides base pair resolution of engaged and active Pol II across the genome. RNA-seq profiles poly-adenylated mRNA transcripts. Created with BioRender.com. b Principal component analysis (PCA) plots showing variation between replicate sets of prophase I substages for leChRO-seq, RNA-seq, and ATAC-seq libraries. c leChRO-seq signal at the Atr locus for LZ (top), P (center), and D (bottom). dREG peaks (teal) are shown for each prophase I substage. TSSs overlapping with dREG peak calls are shaded in blue. Positive values represent the plus strand and negative values represent the minus strand. d Violin plots of pause index values calculated from the ratio of pause peak density to gene body density of leChRO-seq reads on the autosomes. ****p < 0.0001, two-sided Wilcoxon matched-pairs signed rank test. Median value is indicated by the horizontal, center black line. The first and third quartile are indicated by the minima and maxima of the boxplot. n = 4 biologically independent samples. Source data are provided as a Source data file. e Left: Metagene plot of median leChRO-seq signal intensity at annotated gene boundaries. LZ: leptonema/zygonema; pachynema: P; diplonema: D. Right: Inset of leChRO-seq signal for LZ, P, and D at the TSS + 250 bp.