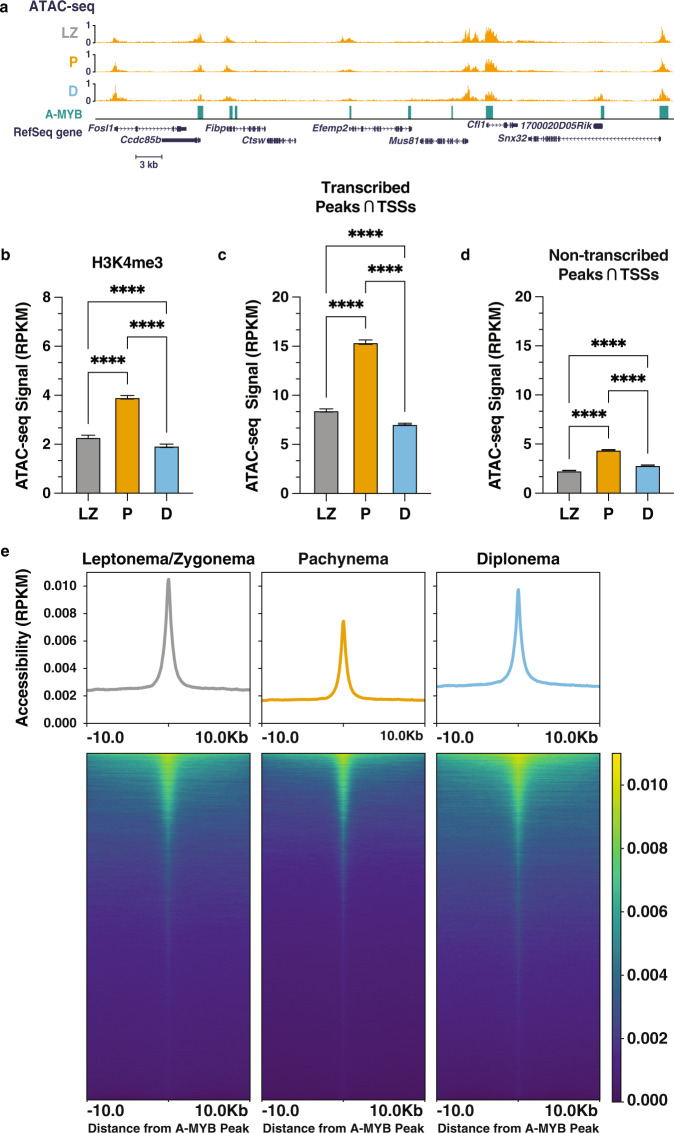
Fig. 6. Reprogramming of meiotic chromatin architecture is facilitated by A-MYB binding.
a Reads per kilobase million (RPKM)-normalized ATAC-seq signal at the Mus81 locus for leptonema/zygonema (top), pachynema (center), and diplonema (bottom). A-MYB ChIP-seq peaks are shown. b–d Stage-resolved RPKM-normalized ATAC-seq signal at H3K4me3 ChIP-seq peaks (b); transcribed ATAC-seq peaks that overlap with annotated transcription start sites (TSSs) (c); non-transcribed ATAC-seq peaks that overlap with TSSs (d). Mean ± 95% confidence intervals are represented on bar graphs. n = 4 biologically independent samples. ****p < 0.0001, two-sided Wilcoxon matched-pairs signed rank test. Source data are provided as a Source data file. e Metaplots (top) and heatmaps (bottom) of RPKM-normalized ATAC-seq signal ± 10 kb from the center of A-MYB ChIP-seq peaks for leptonema/zygonema, pachynema, and diplonema. Peaks are sorted in all prophase I substages by decreasing order of ATAC-seq signal intensity in pachynema.