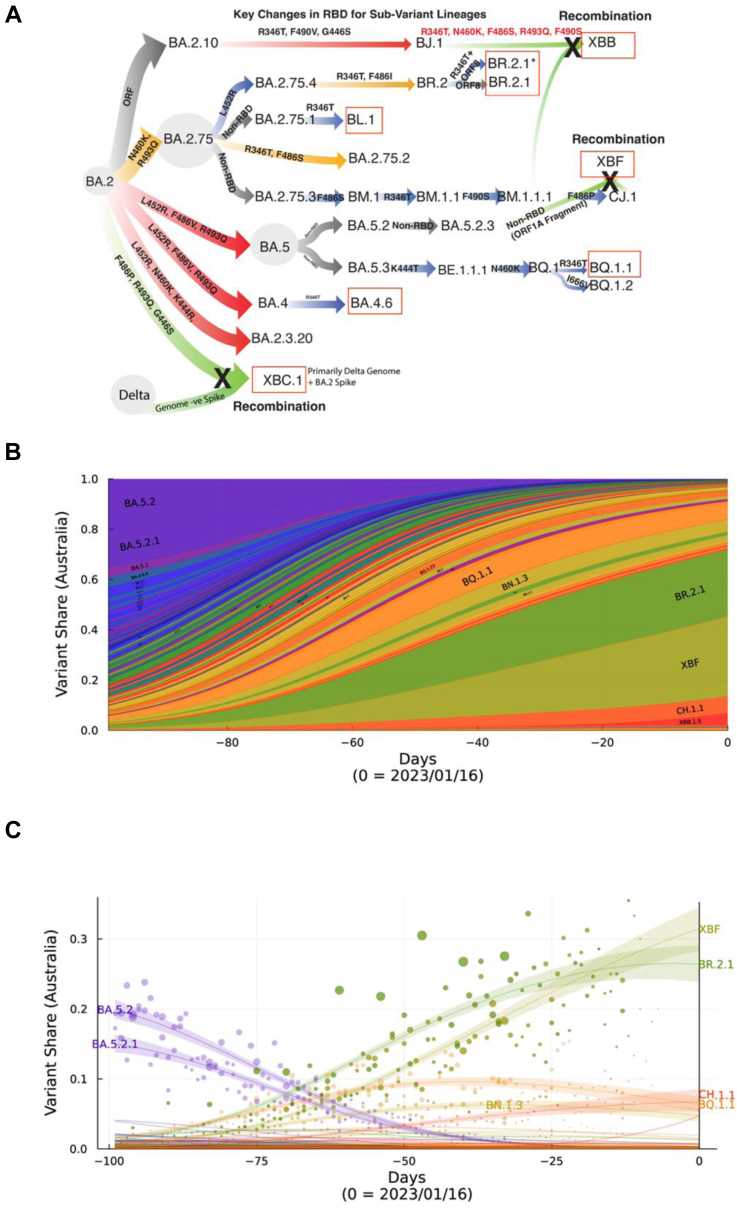
Fig. 1.
Emergence and prevelance of BA.2.75 and BA.5 sub-lineages with convergent Spike polymorphisms in Australia. A. From left to right, Omicron lineages from the parent BA.2 are now primarily split across BA.2.75 and BA.5 sub-lineages. Divergent lineages such as BR.2.1 have accumulated polymorphisms common to BA.5, including L452R and F486I. Whilst BQ.1.1 has acquired BA.2.75 polymorphisms K444T and N460K. Common across most lineages is the additional acquisition of R346T, and this change defines lineages BL.1 and BA.4.6. XBB.1 is a recombinant between BJ.1 and BM.1.1.1 and has benefited from many Spike changes through this recombination event. Whilst XBF is a recombinant between BA.5.2.3 and CJ.1, it retains the latter Spike defined by the F486P change. XBC.1 is a Delta-BA.2 recombinant, with the retention of an intact BA.2 Spike with F486P, G446S and R493Q. Boxed in red are variants that were selected for testing in this study and range from variants with few spike polymorphisms (BL.1 and BA.4.6) through those with many convergent polymorphisms (BR.2.1, XBF and BQ.1.1). For the latter, we have also looked a two circulating isolates of BR.2.1, with and without the R346T change. B. Surveillance summary of Australia at 1601/2 based on genomic data via GISAID. Each vertical slice depicts the posterior mean variant frequency estimate from a hierarchical Bayesian multinomial logistic model of variant competition. Note the two dominant variants BR.2.1 and the recombinant XBF which are dominating in the states of NSW and Victoria, respectively. C. Variant proportions with associated Bayesian Credible Intervals for B.