Abstract
Klebsiella pneumoniae is one of the leading pathogens contributing to antimicrobial resistance. The emergence of carbapenem-resistant K. pneumoniae (CRKP) has put the use of clinical antimicrobial agents in a dilemma. In particular, CRKP exhibiting resistance to ceftazidime/avibactam, tigecycline and colistin have raised great clinical concern, as these are the last-resort antibiotics for the treatment of CRKP infections. Within-host evolution is a survival strategy closely related to the emergence of antimicrobial resistance, while little attention has been paid to the in vivo genetic process of conversion from antibiotic-susceptible to resistant K. pneumoniae. Here we have a literature review regarding the in vivo evolution of resistance to carbapenems, ceftazidime/avibactam, tigecycline, and colistin in K. pneumoniae during antibacterial therapy, and summarized the detailed resistance mechanisms. In general, acquiring blaKPC and blaNDM harboring-plasmid, specific mutations in blaKPC, and porin genes, such as ompK35 and ompK36, upregulation of blaKPC, contribute to the development of carbapenem and ceftazidime/avibactam resistance in vivo. Overexpression of efflux pumps, acquiring plasmid-carrying tet (A) variants, and ribosomal protein change can lead to the adaptive evolution of tigecycline resistance. Specific mutations in chromosomes result in the cationic substitution of the phosphate groups of lipid A, thus contributing to colistin resistance. The resistant plasmid might be acquired from the co-infecting or co-colonizing strains, and the internal environment and antibiotic selection pressure contribute to the emergence of resistant mutants. The internal environment within the human host could serve as an important source of resistant K. pneumoniae strains.
Keywords: Klebsiella pneumoniae, antimicrobial resistance, adaptive evolution, in vivo, antibiotic therapy
Introduction
Klebsiella pneumoniae is a common pathogen for community-and hospital-acquired infections, such as pneumonia, urinary tract infection, and bacteremia (Podschun and Ullmann, 1998; Bengoechea and Sa Pessoa, 2019). The emergence of antimicrobial resistance in K. pneumoniae poses a serious threat to public health, particularly with the rise of carbapenem-resistant K. pneumoniae (CRKP). According to China Antimicrobial Surveillance Network, carbapenem resistance in K. pneumoniae increased rapidly from 3% in 2005 to more than 25% in 2019 (Hu et al., 2020). The irrational use of antibiotics in clinical practice has fostered the occurrence and spread of resistance to “old class antimicrobials.” Ceftazidime/avibactam, tigecycline, and colistin are now considered the last resort for the treatment of CRKP infections (Pournaras et al., 2016; Rabanal and Cajal, 2017; Shields et al., 2017b; Xu et al., 2022). However, the emergence of resistance to the last-resort antibiotics in CRKP has been repeatedly reported (Ah et al., 2014; van Duin et al., 2014; Shields et al., 2016, 2017a). More attention should be paid to the resistance of these types of antimicrobials. Besides, it is nerve-wracking to find that the antimicrobial resistance in K. pneumoniae is changing rapidly in response to in vivo microenvironmental stress during antimicrobial therapy (MacKenzie et al., 1997; Gaibani et al., 2022).
The prevailing view is that the growing prevalence of antimicrobial resistance is largely attributable to selection pressure from antibacterial drugs. However, our understanding of the in vivo development of antimicrobial resistance in K. pneumoniae is limited. While the majority of studies have focused on the epidemiology, risk factors, and treatment outcome of CRKP and other multi-drug resistant K. pneumoniae infections, relatively little attention has been paid to the in vivo genetic process underlying the conversion of a bacterial strain from antibiotic-susceptible to resistant. Within-host evolution is an important survival strategy, often associated with persistent or recurrent infections. Understanding the mechanisms that drive the de novo development of antimicrobial resistance in K. pneumoniae in patients during treatment is crucial for optimizing infection treatment and preventing the emergence of resistance. In this review, we focused on the antimicrobial drugs currently used in clinical practice, and summarized the adaptive evolution of antimicrobial resistance in K. pneumoniae under internal pressures. Our aim is to provide valuable insights into the emergence of antimicrobial resistance in K. pneumoniae during antibacterial therapy.
In vivo adaptive resistance to carbapenem
Acquisition of carbapenemase encoding genes
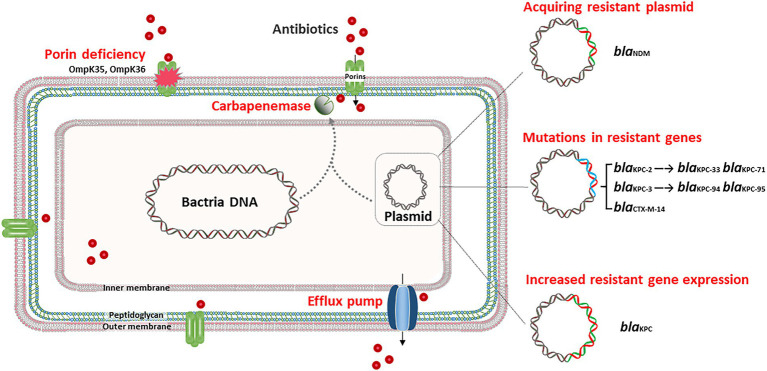
The production of carbapenemase is the leading cause of carbapenem resistance in K. pneumoniae, with K. pneumoniae carbapenemase (KPC) being the most prevalent in several countries, including China (Hu et al., 2012; Giani et al., 2013; Pollett et al., 2014; Zhang et al., 2020). Ding et al. reported that the in vivo acquisition of blaKPC-2 led to carbapenem resistance in K. pneumoniae during antimicrobial therapy, and blaKPC-2 was acquired through horizontal transfer of an insertion sequence containing ISKpn6-like, blaKPC-2 and ISKpn8 (Ding et al., 2016). Duplicative transposition might involve in the mobilization of this insertion sequence, since the transposase gene tnpA was located upstream of blaKPC-2 in KPC plasmids of the CRKP strains. Moreover, in vivo horizontal dissemination of the blaKPC-2 gene carried on IncL/M type conjugative plasmids has been observed among diverse Enterobacteriaceae clinical isolates with different genetic backgrounds, including K. pneumoniae, E. coli, and E. cloacae complex (Anchordoqui et al., 2015). These studies demonstrated that in vivo carbapenem resistance in K. pneumoniae can result from the horizontal transfer of a resistance plasmid or an insertion sequence (Figure 1).
Figure 1.
In vivo adaptive resistance to carbapenem and ceftazidime/avibactam. In general, acquiring blaKPC harboring-plasmid, and specific mutations in porin genes, such as ompK35 and ompK36, contribute to the development of carbapenem resistance in vivo. Acquiring blaNDM harboring-plasmid, specific mutation in blaKPC and upregulation of blaKPC lead to in vivo ceftazidime/avibactam resistance.
Hypervirulent K. pneumoniae (hvKP) can also evolve into carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) under long-term antibiotic treatment by acquiring a resistance plasmid. Recently, a report showed that a sequence type (ST) 218 hvKP strain developed into CR-hvKP in a patient by acquiring an IncFIIk blaKPC-harboring plasmid donated by a ST585 CRKP strain (Tang et al., 2021). K. pneumoniae ST218 is a single-locus variant of ST23 and belongs to clonal group 23, which comprises hypervirulent clones harboring the virulence factors of iuc locus, iro locus, and rmpA/rmpA2 at high frequencies. The IncFIIk blaKPC-harboring plasmid is conjugative and carries five antibiotic resistance genes, blaKPC-2, blaCTX-M-24, tet (A), arr-3, and floR (Table 1).
Table 1.
Examples of studies on in vivo adaptive resistance in Klebsiella pneumoniae.
| Resistant types | Initial strain types | Interval between S and NS isolates (days) | Sequence type (ST) | Underlying mechanisms | Region/Country |
|---|---|---|---|---|---|
| Carbapenem | KP | 23 | 11 | Acquisition of blaKPC-2-harboring Plasmid | Shanghai, China (Ding et al., 2016) |
| 7 | 11 | ||||
| KP | NA | 37 | Acquisition of blaOXA-1-harboring plasmid | Hunan, China (Li et al., 2017) | |
| hvKP | 5 | 218 | Acquisition of blaKPC-harboring plasmid | Hebei, China (Tang et al., 2021) | |
| KP | 14 | 660 | Porin deficiency | Zhejiang, China (Tian et al., 2020) | |
| Ceftazidime/avibactam | KP | 15/14 | 11 | Mutation and reversion of blaKPC genes | Beijing, China (Wang et al., 2021) |
| CRKP | 6 | 11 | Mutations in blaKPC | Henan, China (Li D. et al., 2021) | |
| CRKP | 10 | 101 | Mutations in blaKPC | Italy (Gaibani et al., 2018) | |
| CRKP | 21 | NA | Mutations in blaKPC | Shanghai, China (Shen et al., 2022) | |
| MDR KP | 23 | 383 | Mutations in blaCTX-M-14 | German (Both et al., 2017) | |
| CRKP | 30 | 4496 | blaKPC-2 duplication | Zhejiang, China (Han et al., 2021) | |
| CRKP | 13–22 | 11 | Increased gene expression of mutated blaKPC | Beijing, China (Sun et al., 2021) | |
| CRKP | 11 | 11 | Acquisition of blaNDM-5-harboring plasmid | Fujian, China (Huang et al., 2021) | |
| Tigecycline | CRKP | 41 | 11 | Deletion of the ramR RBS | Shanghai, China (Ye M. et al., 2017) |
| CRKP | 340 | 1544 | Upregulation of RamA and/or RarA | Taiwan, China (Lin et al., 2016) | |
| 37 | 23 | ||||
| 147 | 1526 | ||||
| 7 | 660 | ||||
| 5 | 45 | ||||
| hv-CRKP | 50 | 11 | Mutations of ramR and lon | Zhejiang, China (Jin et al., 2021) | |
| MDR KP | 4 | NA | kpgABC overexpression. | United States (Nielsen et al., 2014) | |
| CRKP | 13 | 11 | The tet(A) variant (S251A) | Zhejiang, China (Du et al., 2018) | |
| CRKP | NA | 11 | Plasmid harbors the blaKPC-2 and tet(A) variant genes | Zhejiang, China (Zhang et al., 2018) | |
| CRKP | 29 | 11 | The rpsJ variant (V57L) | Zhejiang, China (He et al., 2018) | |
| Colistin | CRKP | 30 | 258 | Inactivation or deletion of the mgrB gene | Italy (Cannatelli et al., 2013) |
| CRKP | 12 | 512 | Mutations of pmrB | Italy (Cannatelli et al., 2014) | |
| hv-CRKP | 50 | 11 | Mutations of pmrB, phoQ, and mgrB genes | Zhejiang, China (Jin et al., 2021) |
KP, Klebsiella pneumoniae; CRKP, carbapenem resistant Klebsiella pneumoniae; hvKP, hypervirulent Klebsiella pneumoniae; ST, sequence type; S, susceptible; NS, not susceptible; RBS, ribosomal binding site; MDR, multi-drug resistance; BSI, bloodstream infection; NA, not available.
Porin deficiency
Outer membrane protein (OMP) is the main component of the outer membrane of gram-negative bacteria, including porin and lipoprotein. Porins typically aggregate to form pores, enabling the passage of small hydrophilic molecules, such as β-lactams, across the membrane (Pagès et al., 2008). The porins of K. pneumoniae mainly include OmpK35 and OmpK36 (Hernández-Allés et al., 1999). The deficiency of OmpK35, known as OmpF, is reported widely reported in Enterobacteriaceae strains (Gravey et al., 2020). While OmpK35 is not thought to be the primary pathway for K. pneumoniae resistance (Shields et al., 2015; Pagès et al., 2016). The function of OmpK35 should be further studied in vivo circumstance of K. pneumoniae. OmpK36 is classified as a member of the OmpC porin group, and functions as a non-specific, passive diffusion pore (Ye Y. et al., 2017). The loss of OmpK36, coupled with extended-spectrum β-lactamases (ESBLs) and/or AmpC production, can result in carbapenem resistance in K. pneumoniae (Hamzaoui et al., 2018). Tian et al. (2020) reported three K. pneumoniae strains successively isolated from one patient during hospitalization and found that the final strain developed carbapenem resistance after 14-day of imipenem treatment. This particular CRKP strain exhibited OmpK36 deficiency due to a premature stop codon in the ompK36 gene. This study highlights that the alteration of outer membrane porins due to the 14-day use of imipenem plays a potential role in leading to clinical presentation of carbapenem resistance (Figure 1). In addition, some researchers have gradually discovered that porins are also associated with ceftazidime/avibactam resistance of K. pneumoniae (Xu et al., 2022), which reflects the important role of porins deficiency in the adaptive evolution of K. pneumoniae.
In vivo adaptive resistance to ceftazidime/avibactam
Ceftazidime/avibactam is a novel β-lactam/β-lactamase inhibitor combination, which has been approved for the treatment of complicated intra-abdominal infections and urinary tract infections in 2015 (Zasowski et al., 2015; Shirley, 2018). Avibactam exhibits activity against Ambler class A enzymes (including ESBLs and KPC) and some Ambler class C and D (e.g., OXA-48) enzymes, but it is ineffective against class B enzymes like New Delhi metallo-β-lactamase (NDM) (Shirley, 2018). Though ceftazidime/avibactam displayed potent activity against KPC-producing K. pneumoniae (KPC-KP), the emergence of ceftazidime/avibactam resistance in clinical strains during antimicrobial treatment has been repeatedly reported (Humphries et al., 2015; Shields et al., 2016; Both et al., 2017).
Specific mutations of blaKPC
The major in vivo adaptive ceftazidime/avibactam resistance mechanism is the emergence of specific mutations in blaKPC (Figure 1). A report from China revealed that following a 6-day course of ceftazidime/avibactam treatment, a ST11 K. pneumoniae strain developed ceftazidime/avibactam resistance, owing to the mutation of KPC-2 to KPC-33, with a substitution of D179Y within the Ω loop (Li D. et al., 2021). The Ω loop is located at residues 164–179, which is an essential domain for class A β-lactamases, and the substitution of D179Y may be detrimental to the binding of avibactam (Gaibani et al., 2021; Wang et al., 2021). Another study conducted in Italy showed that in vivo development of ceftazidime/avibactam resistance in K. pneumoniae was also linked to the D179Y substitution in KPC-3, which emerged after 17-day of ceftazidime/avibactam treatment (Gaibani et al., 2018). Similarly, in vivo development of KPC-71, KPC-76, KPC-94, and KPC-95-mediated ceftazidime/avibactam resistance during antimicrobial treatment has also been reported in ST11 and ST512 KPC-KP (Li X. et al., 2021; Guzmán-Puche et al., 2022; Shen et al., 2022). Of note, the low antibiotic pressure may have selected hybrid subpopulations of KPC-KP due to the high adaptability of KPC to ceftazidime/avibactam. For example, a study reported that KPC-KP strains isolated from bronchoalveolar lavage harbor blaKPC-3 and T243M mutations, while those isolated from the blood have D179Y mutation (Gaibani et al., 2021). Except for KPC, the in vivo emerging P170S exchange in CTX-M-14 has also been associated with elevated ceftazidime/avibactam MICs for independent K. pneumoniae isolates, but this substitution was not sufficient for full resistance (Both et al., 2017).
Interestingly, KPC mutations mediating ceftazidime/avibactam resistance are generally associated with the restoration of carbapenem susceptibility (Haidar et al., 2017; Shields et al., 2017a), and this kind of reversion may be dynamic. As illustrated in a recent study, the infection began with a KPC-2-producing K. pneumoniae. After treatment with ceftazidime/avibactam, the strain switched to a KPC-33 mutant (D179Y), which restored carbapenem susceptibility. However, the restored carbapenem susceptibility in vivo was not stable and the subsequent use of imipenem against KPC-33-producing K. pneumoniae infection resulted in a reversion of KPC-2 producers (Wang et al., 2021). The selective pressure of antibiotics in the mutation and reversion of blaKPC genes may lead to the dynamic change of KPC enzymes and the emergence of resistance to ceftazidime/avibactam and carbapenems.
Increased blaKPC gene expression
Increased gene expression and copy number of blaKPC can lead to ceftazidime/avibactam resistance in K. pneumoniae. A study reported that a novel ST4496 strain, which is a novel ST closely related to ST11, displayed ceftazidime/avibactam resistance after 1 month of ceftazidime/avibactam treatment. Sequencing analysis showed that there was duplication of blaKPC-2 on a 108 kb IncFII KPC plasmid due to unequal crossover of the IS26 composite transposon, resulting in elevated levels of blaKPC-2 expression (Han et al., 2021). The increased expression of KPC carbapenemase could lead to enhanced hydrolysis of ceftazidime, as avibactam may not be able to completely inhibit the higher amounts of KPC. Besides, the increased gene expression of blaKPC-51 also leads to high-level ceftazidime/avibactam resistance (Sun et al., 2021).
Acquiring blaNDM-5-harboring plasmid
Acquiring blaNDM-5-harboring plasmid could also lead to ceftazidime/avibactam resistance, since avibactam could not inhibit NDM. One study observed that a ST11 CRKP strain displayed ceftazidime/avibactam resistance after 11-day of treatment. This strain harbored blaKPC-2, blaSHV-182, and blaTEM-1B, and it acquired an additional IncX3 blaNDM-5-harboring plasmid when compared with the corresponding susceptible isolate. This blaNDM-5 plasmid is conjugative and could be successfully transferred into E. coli J53 with high frequency (Huang et al., 2021).
In vivo evolution of tigecycline resistance
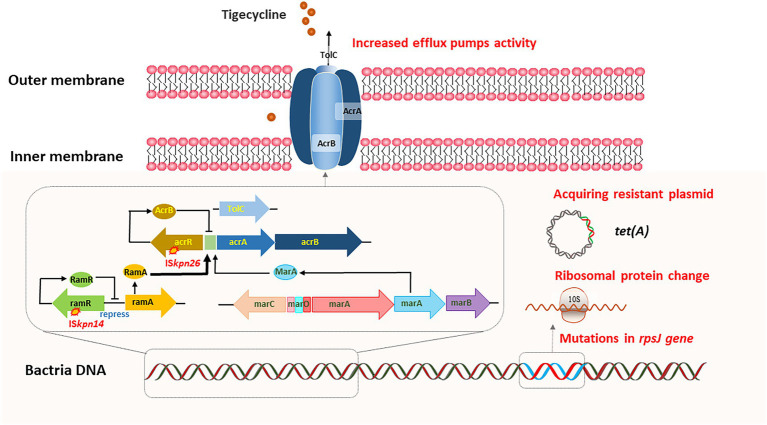
Tigecycline is a derivative of the semi-synthetic tetracycline minocycline, which was approved in China in 2012 (He et al., 2015; Alhashem et al., 2017). It has a broad antibacterial spectrum and displays potent antibacterial activity against gram-positive cocci, gram-negative bacilli (excluding Pseudomonas aeruginosa and some Proteus), anaerobic bacteria, and atypical pathogens (Society of Clinical Microbiology and Infection of China International Exchange and Promotion Association for Medical and Healthcare, Clinical Microbiology Group of the Laboratory Medicine Society of the Chinese Medical Association, Clinical Microbiology G.I.S. ofthe C. M, 2020). Tigecycline mainly binds to the bacterial ribosome 30S subunit, preventing aminoacyl-tRNA from entering the ribosome A site, thus exerting an antibacterial effect by inhibiting bacterial protein synthesis (Velkov et al., 2010). Recently, tigecycline-resistant K. pneumoniae strains have been frequently reported (Sun et al., 2013). Overall, the most commonly reported mechanisms of tigecycline resistance include overexpression of efflux pumps, acquisition of plasmid-carrying tet (A) variants and ribosomal protein change (Figure 2).
Figure 2.
In vivo evolution of tigecycline resistance. Overexpression of efflux pumps, acquiring plasmid-carrying tet(A) variants, and ribosomal protein change can lead to the adaptive evolution of tigecycline resistance.
Increased efflux pumps activity
Efflux pumps can actively squeeze drugs out of cells, and resistance nodulation-cell division (RND) type efflux pump, including AcrAB-TolC and OqxAB, is the dominant drug-associated efflux pump in gram-negative bacteria (Nikaido, 2018). AcrAB-TolC is predominantly related to tigecycline resistance in CRKP (Masi et al., 2019). AcrAB-TolC is mainly composed of three parts: membrane fusion protein (AcrA), efflux transporter (AcrB), and outer membrane channel protein (TolC). Meanwhile, the expression of the AcrAB-TolC is regulated by a variety of regulatory factors, including acrR, ramA, and marA (Wang et al., 2001; Keeney et al., 2007). Studies have shown that bacteria can sense flux rate and regulate efflux pumps to survive the environmental antibiotic challenge, suggesting that resistance mechanisms in vivo may differ from those in vitro (Fritz et al., 2015). Understanding the mechanisms underlying the de novo development of tigecycline resistance in patients is challenging. One strategy is to perform a longitudinal study by isolating K. pneumoniae strains from patients before, during and after tigecycline therapy. Ye et al. reported that the deletion of ramR ribosomal binding site (RBS) could lead to in vivo development of tigecycline resistance in K. pneumoniae after more than 40 days of tigecycline therapy (Ye M. et al., 2017, 2022). RamR exerted negative regulation on acrAB gene expression, and this 12-bp deletion abolished RamR protein production, resulting in high levels of acrAB expression and tigecycline resistance.
In another study, researchers identified five paired clinical isolates of K. pneumoniae that were initially tigecycline-susceptible, but later developed into tigecycline-non-susceptible (Lin et al., 2016). They found that tigecycline-non-susceptibility was associated with upregulation of RamA and/or RarA and the corresponding AcrAB-TolC and/or OqxAB efflux pump (s), respectively. Furthermore, various mutations in ramR and oqxR lead to ramA and rarA overexpression. Meanwhile, AcrAB-TolC efflux pump-mediated tigecycline resistance has also been confirmed in CR-hvKP strains. ST11-KL64 CR-hvKP developed tigecycline resistance due to the mutation of ramR during tigecycline therapy, and a novel frameshift mutation of lon was identified in the high-level tigecycline-resistant strain (Jin et al., 2021). In addition, Nielsen et al. also reported a new efflux pump operon, kpgABC. An insertion sequence (IS5) was correlated with an elevated kpgABC expression, which led to the in vivo development of tigecycline nonsusceptibility in K. pneumoniae (Nielsen et al., 2014).
Acquiring plasmid-carrying tet (A) variants
The widely disseminated plasmid-carrying tet (A) variants in K. pneumoniae have greatly contributed to tigecycline resistance (Chiu et al., 2017). Tet (A) is one of the most common major facilitator superfamilies (MFS) efflux pumps. Tigecycline therapy could upregulate the expression of tet (A) in tigecycline-susceptible CRKP, leading to the development of tigecycline resistance. Du et al. (2018) reported tigecycline-resistant ST11 CRKP isolates from a 56-year-old female patient during tigecycline therapy. One amino acid substitution S251A in TetA was identified in the tigecycline-resistant isolates. Subsequent transformation experiments confirmed the contribution of this TetA variant (S251A) to tigecycline resistance and the tetA gene was located on a transferable plasmid. Another study reported that clinical CRKP strains carrying a conjugative plasmid harboring the blaKPC-2 and tet (A) variant genes readily evolved into tigecycline-resistant CRKP upon treatment and persisted in the human gastrointestinal tract (Zhang et al., 2018).
Ribosomal protein change
Mutations in rpsJ encoding ribosomal protein S10, the target site of tigecycline, have also been associated with tigecycline resistance (Villa et al., 2014). He et al. (2018) monitored a 59-year-old male patient infected with ST11 KPC-producing K. pneumoniae. They identified the V57L amino acid substitution in rpsJ, and confirmed that this mutation was the main cause of tigecycline resistance through transformational complementation assay. This study demonstrated that the evolution of the rpsJ gene could lead to tigecycline resistance in CRKP during tigecycline therapy. Since this gene is located on the chromosome, it provides a clinical warning that under the selective pressure of tigecycline, rpsJ mutations may occur, resulting in drug resistance and treatment failure.
In vivo evolution of colistin resistance
Polymyxins (polymyxin B and colistin) are the key drugs for the treatment of infections caused by CRKP (Livermore et al., 2011; Petrosillo et al., 2013). However, the emergence of colistin-resistant K. pneumoniae has been increasingly reported (Giani et al., 2013; Ah et al., 2014). There are some reports of K. pneumoniae acquiring colistin resistance under in vivo selective pressure.
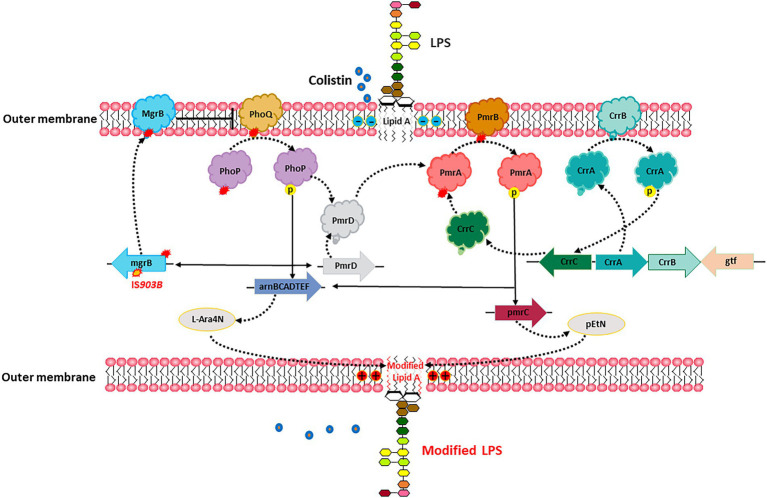
The outer membrane of gram-negative bacteria is the action site of polymyxins due to its affinity for the phosphate group of lipid A. Lipid A is the hydrophobic part located in the outer monolayer of the outer membrane and is synthesized by a series of enzymes encoded by the lpx gene cluster. Lipid A has a negative charge due to the presence of free phosphate groups, while polymyxin has a high affinity for the negative charge of lipid A (Li et al., 2019). As shown in Figure 3, two ionic groups, pEtN and L-Ara4N, can impact the affinity between polymyxin and the outer membrane and lead to resistance. The expression of these two ionic groups is controlled by LPS-modifying enzyme genes pmrC, pmrE, and pmrHFIJKLM, which is then controlled by PmrAB and PhoPQ two-component system (Kox, 2000; Lee et al., 2004; Olaitan et al., 2014). In addition, CrrAB and MgrB are also involved in this regulatory axis. CrrAB could regulate the expression of PmrAB and PhoPQ (Olaitan et al., 2014), while MgrB is a negative regulator of the PhoPQ signaling system. The mutations in certain genes are involved in the regulation of these signaling pathways, such as pmrAB, phoPQ, mgrB, crrB, which can lead to colistin resistance (Cheng et al., 2010; Cannatelli et al., 2013; Chen and Groisman, 2013; Xie et al., 2022).
Figure 3.
In vivo evolution of colistin resistance. Specific mutations in chromosomes led to the cationic substitution of the phosphate groups of lipid A, contributing to colistin resistance.
Comparative genomic analysis of a pair of sequential KPC-KP isolates from the same patient, including a colistin-susceptible isolate and a colistin-resistant isolate selected after colistin exposure, revealed that insertional inactivation of the mgrB gene is a genetic mechanism for acquired colistin resistance (Cannatelli et al., 2013). Another study found a non-synonymous nucleotide substitution in the pmrB gene that resulted in a leucine-to-arginine substitution at amino acid position 82 in CRKP-infected patients treated with low-dose colistin (Cannatelli et al., 2014). This substation upregulated transcription of pmrA and pmrK, which is part of the pmrHFIJKLM operon responsible for modification of the colistin lipopolysaccharide target. Besides, the mutations of pmrB, phoQ and mgrB genes also account for the in-host evolution of CR-hvKP to colistin resistance (Jin et al., 2021).
Conclusion
Our work summarized in vivo adaptive evolution of antimicrobial resistance in K. pneumoniae during antimicrobial therapy in currently clinical practice. We further described the underlying mechanisms of evolved resistance to carbapenems, ceftazidime/avibactam, tigecycline, and colistin within human hosts. In general, acquiring blaKPC and blaNDM harboring-plasmid, specific mutations in blaKPC, and porin genes, ompK35 and ompK36, upregulation of blaKPC, contribute to the development of carbapenems and ceftazidime/avibactam resistance in vivo. Overexpression of efflux pumps, acquiring plasmid-carrying tet (A) variants, and ribosomal protein change can lead to the adaptive evolution of tigecycline resistance. Specific mutations in chromosomes result in a variety of modifications of LPS, contributing to colistin resistance.
The adaptive evolution of K. pneumoniae can be attributed to the impact of the human host’s internal environment and antibiotic selection pressure. The in vivo development of antimicrobial resistance in K. pneumoniae was established mainly through the acquisition of a resistant plasmid and the emergence of specific mutations. Plasmid-carried resistant genes have been proven to disseminate ubiquitously, such as blaKPC and tet (A). Moreover, acquired antimicrobial resistance in hvKP clones demonstrated that in vivo adaptive evolution also promotes the convergence of hypervirulence and resistance.
The resistant plasmid might be acquired from the co-infecting or co-colonizing strains, and the internal selection factors may contribute to the horizontal transfer of plasmid within the human host. The emerging resistant K. pneumoniae strains in vivo could then disseminate through nosocomial settings and be screened by sectional molecular epidemiology studies. It is possible that the internal environment within the human host could serve as an important source of resistant K. pneumoniae strains. In the future, more attention should be paid to the in vivo genetic process of conversion from antibiotic-susceptible to resistant K. pneumoniae, and the high possibility of convergence of hypervirulence and resistance. The evolution of resistant strains could be effectively reduced by blocking this conversion process.
Author contributions
SL and ML: conceptualization. SL and XF: methodology and writing-original draft preparation. ZS: validation and supervision. SL and ZS: writing-review and editing. ML: project administration. SL: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ningbo Medical Science and Technology Project, China (Grant no. 2021Y83), the National Natural Science Foundation of China (82272374), and the Ningbo Hangzhou Bay Hospital Qihang Talent Program (Grant no. WY-KY-QH-2021-003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Ah Y.-M., Kim A.-J., Lee J.-Y. (2014). Colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 44, 8–15. doi: 10.1016/j.ijantimicag.2014.02.016 [DOI] [PubMed] [Google Scholar]
- Alhashem F., Tiren-Verbeet N. L., Alp E., Doganay M. (2017). Treatment of sepsis: what is the antibiotic choice in bacteremia due to carbapenem resistant Enterobacteriaceae? World J. Clin. Cases 5, 324–332. doi: 10.12998/wjcc.v5.i8.324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchordoqui M. S., De Belder D., Lucero C., Rapoport M., Faccone D., Rodriguez A., et al. (2015). In vivo horizontal dissemination of the blaKPC-2 gene carried on diverse genetic platforms among clinical isolates of Enterobacteriaceae. J. Glob. Antimicrob. Resist. 3, 210–213. doi: 10.1016/j.jgar.2015.05.001, PMID: [DOI] [PubMed] [Google Scholar]
- Bengoechea J. A., Sa Pessoa J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43, 123–144. doi: 10.1093/femsre/fuy043, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both A., Büttner H., Huang J., Perbandt M., Belmar Campos C., Christner M., et al. (2017). Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 72, 2483–2488. doi: 10.1093/jac/dkx179, PMID: [DOI] [PubMed] [Google Scholar]
- Cannatelli A., D’Andrea M. M., Giani T., Di Pilato V., Arena F., Ambretti S., et al. (2013). In vivo emergence of Colistin resistance in Klebsiella pneumoniae producing KPC-type Carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A., Di Pilato V., Giani T., Arena F., Ambretti S., Gaibani P., et al. (2014). In vivo evolution to Colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 58, 4399–4403. doi: 10.1128/AAC.02555-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. D., Groisman E. A. (2013). The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67, 83–112. doi: 10.1146/annurev-micro-092412-155751, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.-Y., Chen Y.-F., Peng H.-L. (2010). Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 17:60. doi: 10.1186/1423-0127-17-60, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.-K., Huang L.-Y., Chen H., Tsai Y.-K., Liou C.-H., Lin J.-C., et al. (2017). Roles of ramR and tet (a) mutations in conferring Tigecycline resistance in Carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 61:e00391-17. doi: 10.1128/AAC.00391-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Shen Z., Hu F., Ye M., Xu X., Guo Q., et al. (2016). In vivo acquisition of carbapenemase gene blakpc-2 in multiple species of enterobacteriaceae through horizontal transfer of insertion sequence or plasmid. Front. Microbiol. 7, 1–8. doi: 10.3389/fmicb.2016.01651, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., He F., Shi Q., Zhao F., Xu J., Fu Y., et al. (2018). The rapid emergence of tigecycline resistance in blaKPC-2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front. Microbiol. 9, 1–7. doi: 10.3389/fmicb.2018.00648, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz G., Dintner S., Treichel N. S., Radeck J., Gerland U., Mascher T., et al. (2015). A new way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. MBio 6:e00975. doi: 10.1128/mBio.00975-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani P., Bovo F., Bussini L., Lazzarotto T., Amadesi S., Bartoletti M., et al. (2022). Dynamic evolution of imipenem/relebactam resistance in a KPC-producing Klebsiella pneumoniae from a single patient during ceftazidime/avibactam-based treatments. J. Antimicrob. Chemother. 77, 1570–1577. doi: 10.1093/jac/dkac100, PMID: [DOI] [PubMed] [Google Scholar]
- Gaibani P., Campoli C., Lewis R. E., Volpe S. L., Scaltriti E., Giannella M., et al. (2018). In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J. Antimicrob. Chemother. 73, 1525–1529. doi: 10.1093/jac/dky082, PMID: [DOI] [PubMed] [Google Scholar]
- Gaibani P., Gatti M., Rinaldi M., Crovara Pesce C., Lazzarotto T., Giannella M., et al. (2021). Suboptimal drug exposure leads to selection of different subpopulations of ceftazidime-avibactam-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in a critically ill patient. Int. J. Infect. Dis. 113, 213–217. doi: 10.1016/j.ijid.2021.10.028, PMID: [DOI] [PubMed] [Google Scholar]
- Giani T., Pini B., Arena F., Conte V., Bracco S., Migliavacca R., et al. (2013). Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 may to 30 June 2011. Euro Surveill. 18:20489. [PubMed] [Google Scholar]
- Gravey F., Cattoir V., Ethuin F., Fabre L., Beyrouthy R., Bonnet R., et al. (2020). ramR deletion in an Enterobacter hormaechei isolate as a consequence of therapeutic failure of key antibiotics in a long-term hospitalized patient. Antimicrob. Agents Chemother. 64:e00962-20. doi: 10.1128/AAC.00962-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán-Puche J., Pérez-Nadales E., Pérez-Vázquez M., Causse M., Gracia-Ahufinger I., Mendez-Natera A., et al. (2022). In vivo selection of KPC-94 and KPC-95 in Klebsiella pneumoniae isolates from patients treated with ceftazidime/avibactam. Int. J. Antimicrob. Agents 59:106524. doi: 10.1016/j.ijantimicag.2022.106524, PMID: [DOI] [PubMed] [Google Scholar]
- Haidar G., Clancy C. J., Shields R. K., Hao B., Cheng S., Nguyen M. H. (2017). Mutations in Bla KPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-Spectrum β-lactamases. Antimicrob. Agents Chemother. 61:e02534-16. doi: 10.1128/AAC.02534-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzaoui Z., Ocampo-Sosa A., Fernandez Martinez M., Landolsi S., Ferjani S., Maamar E., et al. (2018). Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int. J. Antimicrob. Agents 52, 898–905. doi: 10.1016/j.ijantimicag.2018.03.020, PMID: [DOI] [PubMed] [Google Scholar]
- Han X., Shi Q., Mao Y., Quan J., Zhang P., Lan P., et al. (2021). Emergence of ceftazidime/avibactam and Tigecycline resistance in Carbapenem-resistant Klebsiella pneumoniae due to in-host microevolution. Front. Cell. Infect. Microbiol. 11, 1–9. doi: 10.3389/fcimb.2021.757470, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Fu Y., Chen Q., Ruan Z., Hua X., Zhou H., et al. (2015). Tigecycline susceptibility and the role of efflux pumps in Tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One 10:e0119064. doi: 10.1371/journal.pone.0119064, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Shi Q., Fu Y., Xu J., Yu Y., Du X. (2018). Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect. Genet. Evol. 66, 188–191. doi: 10.1016/j.meegid.2018.09.025 [DOI] [PubMed] [Google Scholar]
- Hernández-Allés S., Albertí S., Álvarez D., Doménech-Sánchez A., Martínez-Martínez L., Gil J., et al. (1999). Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145, 673–679. doi: 10.1099/13500872-145-3-673 [DOI] [PubMed] [Google Scholar]
- Hu F., Chen S., Xu X., Guo Y., Liu Y., Zhu D., et al. (2012). Emergence of carbapenem-resistant clinical Enterobacteriaceae isolates from a teaching hospital in Shanghai. China. J. Med. Microbiol. 61, 132–136. doi: 10.1099/jmm.0.036483-0, PMID: [DOI] [PubMed] [Google Scholar]
- Hu F., Guo Y., Zhu D. (2020). CHINET surveillance of bacterial resistance across tertiary hospitals in 2019. Chin J infect Chemother. Chin. J. Infect. Chemother. 20, 233–243. [Google Scholar]
- Huang J., Zhang S., Zhao Z., Chen M., Cao Y., Li B. (2021). Acquisition of a Stable and Transferable Bla NDM-5-positive plasmid with low fitness cost leading to ceftazidime/avibactam resistance in KPC-2-producing Klebsiella pneumoniae during treatment. Front. Cell. Infect. Microbiol. 11, 1–8. doi: 10.3389/fcimb.2021.658070, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries R. M., Yang S., Hemarajata P., Ward K. W., Hindler J. A., Miller S. A., et al. (2015). First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 59, 6605–6607. doi: 10.1128/AAC.01165-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Chen Q., Shen F., Jiang Y., Wu X., Hua X., et al. (2021). Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg. Microbes Infect. 10, 1129–1136. doi: 10.1080/22221751.2021.1937327, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney D., Ruzin A., McAleese F., Murphy E., Bradford P. A. (2007). MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J. Antimicrob. Chemother. 61, 46–53. doi: 10.1093/jac/dkm397, PMID: [DOI] [PubMed] [Google Scholar]
- Kox L. F. F. (2000). A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19, 1861–1872. doi: 10.1093/emboj/19.8.1861, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Hsu F.-F., Turk J., Groisman E. A. (2004). The PmrA-regulated pmrC gene mediates Phosphoethanolamine modification of lipid a and Polymyxin resistance in salmonella enterica. J. Bacteriol. 186, 4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ke H., Wu W., Tu Y., Zhou H., Yu Y. (2021). Molecular mechanisms driving the in vivo development of KPC-71-mediated resistance to ceftazidime-avibactam during treatment of Carbapenem-resistant Klebsiella pneumoniae infections. mSphere 6:e0085921. doi: 10.1128/mSphere.00859-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Li K., Dong H., Ren D., Gong D., Jiang F., et al. (2021). Ceftazidime-avibactam resistance in klebsiella pneumoniae sequence type 11 due to a mutation in plasmid-borne blakpc-2 to blakpc-33, in Henan. China. Infect. Drug Resist. 14, 1725–1731. doi: 10.2147/IDR.S306095, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nation R. L., Kaye K. S. (2019). Polymyxin antibiotics: from laboratory bench to bedside. Berlin: Springer International Publishing. [Google Scholar]
- Li P., Wang M., Li X., Hu F., Yang M., Xie Y., et al. (2017). ST37 Klebsiella pneumoniae: development of carbapenem resistance in vivo during antimicrobial therapy in neonates. Future Microbiol. 12, 891–904. doi: 10.2217/fmb-2016-0165 [DOI] [PubMed] [Google Scholar]
- Lin Y. T., Huang Y. W., Huang H. H., Yang T. C., Wang F., Fung C. P. (2016). In vivo evolution of tigecycline-non-susceptible Klebsiella pneumoniae strains in patients: relationship between virulence and resistance. Int. J. Antimicrob. Agents 48, 485–491. doi: 10.1016/j.ijantimicag.2016.07.008, PMID: [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Warner M., Mushtaq S., Doumith M., Zhang J., Woodford N. (2011). What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int. J. Antimicrob. Agents 37, 415–419. doi: 10.1016/j.ijantimicag.2011.01.012, PMID: [DOI] [PubMed] [Google Scholar]
- MacKenzie F., Forbes K., Dorai-John T., Amyes S., Gould I. (1997). Emergence of a carbapenem-resistant Kiebsiella pneumoniae. Lancet 350:783. doi: 10.1016/S0140-6736(05)62567-6 [DOI] [PubMed] [Google Scholar]
- Masi M., Winterhalter M., Pagès J.-M. (2019). “Outer membrane porins” in Bacterial cell walls and membranes. ed. Kuhn A. (Berlin: Springer International Publishing; ), 79–123. [Google Scholar]
- Nielsen L. E., Snesrud E. C., Onmus-Leone F., Kwak Y. I., Avilés R., Steele E. D., et al. (2014). IS 5 element integration, a novel mechanism for rapid in vivo emergence of Tigecycline nonsusceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 58, 6151–6156. doi: 10.1128/AAC.03053-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2018). RND transporters in the living world. Res. Microbiol. 169, 363–371. doi: 10.1016/j.resmic.2018.03.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J.-M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:e00643. doi: 10.3389/fmicb.2014.00643, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès J.-M., James C. E., Winterhalter M. (2008). The porin and the permeating antibiotic: a selective diffusion barrier in gram-negative bacteria. Nat. Rev. Microbiol. 6, 893–903. doi: 10.1038/nrmicro1994, PMID: [DOI] [PubMed] [Google Scholar]
- Pagès J.-M., Peslier S., Keating T. A., Lavigne J.-P., Nichols W. W. (2016). Role of the outer membrane and Porins in susceptibility of β-lactamase-producing Enterobacteriaceae to ceftazidime-avibactam. Antimicrob. Agents Chemother. 60, 1349–1359. doi: 10.1128/AAC.01585-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Giannella M., Lewis R., Viale P. (2013). Treatment of carbapenem-resistant Klebsiella pneumoniae: the state of the art. Expert Rev. Anti-Infect. Ther. 11, 159–177. doi: 10.1586/eri.12.162, PMID: [DOI] [PubMed] [Google Scholar]
- Podschun R., Ullmann U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollett S., Miller S., Hindler J., Uslan D., Carvalho M., Humphries R. M. (2014). Phenotypic and molecular characteristics of Carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J. Clin. Microbiol. 52, 4003–4009. doi: 10.1128/JCM.01397-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras S., Koumaki V., Spanakis N., Gennimata V., Tsakris A. (2016). Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int. J. Antimicrob. Agents 48, 11–18. doi: 10.1016/j.ijantimicag.2016.04.017, PMID: [DOI] [PubMed] [Google Scholar]
- Rabanal F., Cajal Y. (2017). Recent advances and perspectives in the design and development of polymyxins. Nat. Prod. Rep. 34, 886–908. doi: 10.1039/C7NP00023E, PMID: [DOI] [PubMed] [Google Scholar]
- Shen S., Shi Q., Hu F. (2022). The changing face of Klebsiella pneumoniae carbapenemase: in-vivo mutation in patient with chest infection. Lancet 399:2226. doi: 10.1016/S0140-6736(22)01011-X, PMID: [DOI] [PubMed] [Google Scholar]
- Shields R. K., Chen L., Cheng S., Chavda K. D., Press E. G., Snyder A., et al. (2017a). Emergence of ceftazidime-avibactam resistance due to plasmid-borne Bla KPC-3 mutations during treatment of Carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob. Agents Chemother. 61:e02097-16. doi: 10.1128/AAC.02097-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. K., Clancy C. J., Hao B., Chen L., Press E. G., Iovine N. M., et al. (2015). Effects of Klebsiella pneumoniae Carbapenemase subtypes, extended-Spectrum β-lactamases, and Porin mutations on the in vitro activity of ceftazidime-avibactam against Carbapenem-Resistant K. pneumoniae. Antimicrob. Agents Chemother. 59, 5793–5797. doi: 10.1128/AAC.00548-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. K., Nguyen M. H., Chen L., Press E. G., Potoski B. A., Marini R. V., et al. (2017b). Ceftazidime-avibactam is superior to other treatment regimens against Carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrob. Agents Chemother. 61:e00883-17. doi: 10.1128/AAC.00883-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. K., Potoski B. A., Haidar G., Hao B., Doi Y., Chen L., et al. (2016). Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for Carbapenem-resistant Enterobacteriaceae infections. Clin. Infect. Dis. 63, 1615–1618. doi: 10.1093/cid/ciw636, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley M. (2018). Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs 78, 675–692. doi: 10.1007/s40265-018-0902-x, PMID: [DOI] [PubMed] [Google Scholar]
- Society of Clinical Microbiology and Infection of China International Exchange and Promotion Association for Medical and Healthcare, Clinical Microbiology Group of the Laboratory Medicine Society of the Chinese Medical Association, Clinical Microbiology G.I.S. ofthe C. M (2020). Expert consensus on polymyxins, tigecycline and ceftazidime/avibactam susceptibility testing. Chinese J. Lab. Med. 43, 964–972. doi: 10.3760/cma.j.cn114452-20200719-00619 [DOI] [Google Scholar]
- Sun Y., Cai Y., Liu X., Bai N., Liang B., Wang R. (2013). The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents 41, 110–116. doi: 10.1016/j.ijantimicag.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Sun L., Li H., Wang Q., Liu Y., Cao B. (2021). Increased gene expression and copy number of mutated Bla KPC lead to high-level ceftazidime/avibactam resistance in Klebsiella pneumoniae. BMC Microbiol. 21, 1–10. doi: 10.1186/s12866-021-02293-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Hu J., Zhao Y., Song Y., Wang C., Zhang G., et al. (2021). In vivo evolution of carbapenem resistance in hypervirulent Klebsiella pneumoniae in a patient undergoing long-term treatment. J. Antimicrob. Chemother. 77, 531–533. doi: 10.1093/jac/dkab380 [DOI] [PubMed] [Google Scholar]
- Tian X., Wang Q., Perlaza-Jiménez L., Zheng X., Zhao Y., Dhanasekaran V., et al. (2020). First description of antimicrobial resistance in carbapenem-susceptible Klebsiella pneumoniae after imipenem treatment, driven by outer membrane remodeling. BMC Microbiol. 20, 218–211. doi: 10.1186/s12866-020-01898-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D., Cober E. D., Richter S. S., Perez F., Cline M., Kaye K. S., et al. (2014). Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin. Microbiol. Infect. 20, O1117–O1120. doi: 10.1111/1469-0691.12714, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T., Thompson P. E., Nation R. L., Li J. (2010). Structure−activity relationships of Polymyxin antibiotics. J. Med. Chem. 53, 1898–1916. doi: 10.1021/jm900999h, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Feudi C., Fortini D., García-Fernández A., Carattoli A. (2014). Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring Tigecycline resistance. Antimicrob. Agents Chemother. 58, 1707–1712. doi: 10.1128/AAC.01803-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Dzink-Fox J. L., Chen M., Levy S. B. (2001). Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45, 1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhao J., Liu Z., Sun A., Sun L., Li B., et al. (2021). In vivo selection of imipenem resistance among ceftazidime-avibactam-resistant, imipenem-susceptible Klebsiella pneumoniae isolate with KPC-33 Carbapenemase. Front. Microbiol. 12:e727946. doi: 10.3389/fmicb.2021.727946, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Chen K., Dong N., Xu Q., Chan E. W.-C., Zhang R., et al. (2022). Phenotypic changes associated with in vivo evolution of Colistin resistance in ST11 Carbapenem-resistant Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 12:e841748. doi: 10.3389/fcimb.2022.841748, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Guo Y., Ji Y., Wang B., Zhou K. (2022). Epidemiology and mechanisms of Ceftazidime–Avibactam resistance in gram-negative bacteria. Engineering 11, 138–145. doi: 10.1016/j.eng.2020.11.004 [DOI] [Google Scholar]
- Ye M., Ding B., Qian H., Xu Q., Jiang J., Huang J., et al. (2017). In vivo development of tigecycline resistance in Klebsiella pneumoniae owing to deletion of the ramR ribosomal binding site. Int. J. Antimicrob. Agents 50, 523–528. doi: 10.1016/j.ijantimicag.2017.04.024, PMID: [DOI] [PubMed] [Google Scholar]
- Ye M., Liao C., Shang M., Zou D., Feng X., Lu X., et al. (2022). Reduced virulence and enhanced host adaption during antibiotics therapy: a story of a within-host Carbapenem-resistant Klebsiella pneumoniae sequence type 11 evolution in a patient with a serious scrotal abscess. mSystems 7:e0134221. doi: 10.1128/msystems.01342-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Xu L., Han Y., Chen Z., Liu C., Ming L. (2017). Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp. Ther. Med. 15, 1143–1149. doi: 10.3892/etm.2017.5485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasowski E. J., Rybak J. M., Rybak M. J. (2015). The β-lactams strike Back: ceftazidime-avibactam. Pharmacother. J. Hum. Pharmacol. Drug Ther. 35, 755–770. doi: 10.1002/phar.1622, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Dong N., Huang Y., Zhou H., Xie M., Chan E. W. C., et al. (2018). Evolution of tigecycline-and colistin-resistant CRKP (carbapenem-resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract article. Emerg. Microbes Infect. 7, 1–11. doi: 10.1038/s41426-018-0129-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Shi Q., Hu H., Hong B., Wu X., Du X., et al. (2020). Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 26, 124.e1–124.e4. doi: 10.1016/j.cmi.2019.08.020 [DOI] [PubMed] [Google Scholar]