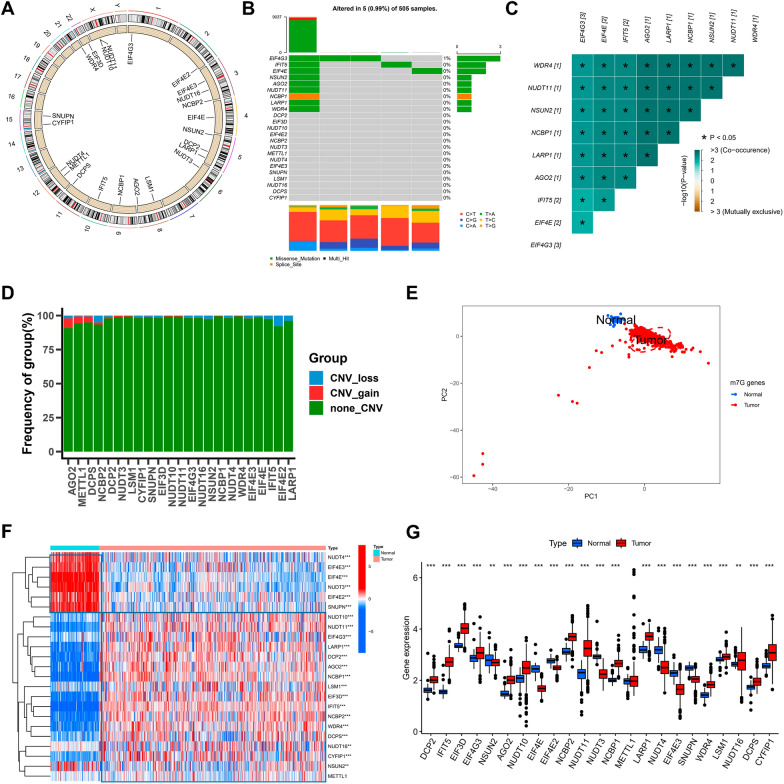
Fig. 1.
Landscape of m7G regulators in LGG. A The 23 chromosomal regions where m7G regulator CNV alterations were found in TCGA-LGG cohorts. B The frequency of m7G regulator mutations and their categorization. C Using Pearson correlation analysis, a correlation plot drawn between the top 9 m7G regulator mutation frequencies. p < 0.05 was considered significant. D The m7G regulators’ CNV variation frequency in the TCGA-LGG cohort. E The m7G regulators-transcriptome profiles of healthy and malignant tissue were analyzed using principal component analysis, which revealed a remarkable difference between the various samples. F The m7G regulators’ expression varied between healthy and malignant tissues, as shown by a heatmap. G The 23 m7G regulators’ expression in LGG tissue from the TCGA-LGG cohort versus healthy tissues from Genotype-Tissue Expression samples. The Kruskal–Wallis test was used to compare the difference. * p < 0.05; ** p < 0.01; *** p < 0.001