Abstract
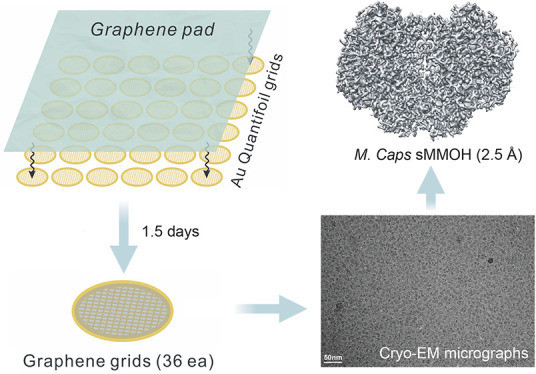
Cryogenic electron microscopy (cryo-EM) has become a widely used tool for determining the protein structure. Despite recent technical advances, sample preparation remains a major bottleneck for several reasons, including protein denaturation at the air–water interface, the presence of preferred orientations, nonuniform ice layers, etc. Graphene, a two-dimensional allotrope of carbon consisting of a single atomic layer, has recently gained attention as a near-ideal support film for cryo-EM that can overcome these challenges because of its superior properties, including mechanical strength and electrical conductivity. Here, we introduce a reliable, easily implemented, and reproducible method to produce 36 graphene-coated grids within 1.5 days. To demonstrate their practical application, we determined the cryo-EM structure of Methylococcus capsulatus soluble methane monooxygenase hydroxylase (sMMOH) at resolutions of 2.9 and 2.5 Å using Quantifoil and graphene-coated grids, respectively. We found that the graphene-coated grid has several advantages, including a smaller amount of protein required and avoiding protein denaturation at the air–water interface. By comparing the cryo-EM structure of sMMOH with its crystal structure, we identified subtle yet significant geometrical changes at the nonheme diiron center, which may better indicate the active site configuration of sMMOH in the resting/oxidized state.
Keywords: Cryogenic electron microscopy, Single particle analysis, Graphene grid, M. caps sMMOH, Tomography
Introduction
Single-particle cryogenic electron microscopy (cryo-EM) has evolved as a major technique to determine the high-resolution protein structure. Advances in multidisciplinary technologies including direct electron detectors, energy filter systems, advanced algorithms, and data collection/processing strategies have enabled cryo-EM to become a versatile and routine method for the structure determination of biomacromolecules.1−6
With these advances and the broad adoption of cryo-EM techniques in instrument development and software algorithms, specimen preparation and grid development have become increasingly important for determining protein structures by cryo-EM.7−9 Several challenges remain in cryo-EM sample preparation, such as protein denaturation mediated by the air–water interface (AWI), nonuniform ice thickness, preferred particle distributions/orientations, and beam-induced motion.10−15 To overcome these challenges, the addition of a continuous thin layer of supporting film on the cryo-EM grid has been widely considered and tested; film materials include inorganic metal alloys (e.g., titanium–silicon and nickel–titanium), carbon nanomembranes, and other forms of amorphous carbon.16−20 However, these films typically add significant background noise to the microscopic images. Other types of supporting films, such as a lipid monolayer and two-dimensional (2D) streptavidin crystals, have been successfully employed to determine protein structures;21−25 however, technical challenges have limited their availability and applicability.
An ideal supporting film for cryo-EM grids must have several properties. First, the material must be thin and sufficiently transparent so that it does not interrupt the electron beam pathway to minimize unwanted scattering. Second, it should be physically strong enough to stably hold both a thin ice layer and particles during the screening and data collection. Finally, it should be electrically conductive to prevent charge accumulation on the surface during the lengthy automated data collection. Graphene, a 2D single atomic carbon layer, meets most of the conditions for use as a supporting film, as it possesses electrical conductivity (∼15,000 cm2 V–1 s–1), optical transparency (∼97.7%), mechanical strength (∼1000 GPa), and minimal scattering events under a 300 kV electron beam.26−30 A plasma-treated graphene grid has been shown to achieve more evenly distributed particles in ice and to minimize beam-induced particle motion.31
Several research groups recently attempted to coat EM grids with graphene or its derivatives to exploit the superior properties for cryo-EM observation.14,25,32−38 Although these earlier attempts were successful, the methods they used are not easily adopted at the laboratory level for several reasons. (1) Some of the reported methods require professional instruments, such as chemical vapor deposition (CVD) equipment for graphene synthesis and a Langmuir–Blodgett trough for uniform graphene oxide coating.38,39 In addition, it is necessary to collaborate with an expert in graphene or its derivatives during its synthesis, transfer, and modification. (2) Only a few established methods are available for assessing the quality of graphene-coated grids.33,35,40
Here, we report a versatile and easy fabrication method for developing graphene-coated grids. We focused on developing a robust graphene-coating protocol for grids that is reproducible and easy to follow. Using our method, we routinely prepared 36 graphene-coated grids at once in ∼1.5 days. We also focused on establishing proper validation methods to evaluate the success of the graphene-coating process on a grid.
To test the applicability and quality of the graphene-coated grids, we determined the cryo-EM structures of Methylococcus capsulatus (M. caps) soluble methane monooxygenase hydroxylase (sMMOH) using the recently developed graphene-coated grid. Methanotrophic bacteria in ambient environments use methane as their sole carbon source by converting methane to methanol. The critical enzymes participating in this conversion are two types of methane monooxygenases: particulate methane monooxygenase (pMMO) and soluble methane monooxygenase (sMMO). In particular, sMMO participates in the following reaction:
The effectiveness of catalysis by sMMO depends on the interplay of four protein components: sMMO hydroxylase (sMMOH), sMMO reductase (sMMOR), sMMO regulatory subunit (sMMOB), and sMMO inhibitory subunit (sMMOD). The crystal structure of sMMOH exhibits a 251 kDa heterohexameric (α2β2γ2) architecture with a glutamate- and histidine-coordinated diiron active site in each α protomer.41 sMMOR shuttles electrons from reduced nicotinamide adenine dinucleotide (NADH) to the diiron center of sMMOH.42 sMMOB is an auxiliary component that modulates the reduction potentials at the diiron center and facilitates substrate availability at the active site.43,44 Another regulatory component, sMMOD, inhibits the catalytic activity of sMMOH.45,46 To date, all 28 structures of sMMOH deposited since 1993 have been determined solely by X-ray crystallography.41,47−49 However, X-ray structures can be affected by high precipitant concentrations and crystal packing. To obtain the atomic-resolution structure without unnatural compounds and crystal contacts, we determined the cryo-EM structure of M. caps sMMOH at resolutions of 2.9 and 2.5 Å using Au Quantifoil and the recently developed graphene-coated grid, respectively.
Results and Discussion
Making Graphene-Coated Au Quantifoil Grids
The graphene transfer approach we developed and adopted is based on the polymer-film-assisted transfer method instead of the polymer-free transfer method.50−52 Although the polymer-free transfer method has several advantages (for example, it can produce a hyperclean graphene surface and graphene with high-order crystallinity), skills and knowledge are required for interfacial control between the transfer solvents, and thus the method is not suitable for those who are unfamiliar with the handling of graphene and surface engineering. By contrast, polymer-assisted graphene transfer has several advantages that make it possible to overcome these challenges. (1) It provides an intuitive and easy-to-use method of generating graphene-coated Au Quantifoil grids (i.e., graphene grids) at the laboratory level for those who are unfamiliar with nanomaterials. (2) It can be used for the mass production of high-quality graphene grids with good surface coverage (>95%) and cleanness. It takes 1.5 days to generate a batch of 36 graphene grids using the method we developed. The method requires a Petri dish, an oven, a 25 × 25 mm PMMA/graphene pad (ACS Material), acetone, and a 3D-printed graphene transfer tool that we designed and developed (Figure S1). The graphene grid synthesis process is described in detail in Experimental Methods section (Movie S1) and illustrated in Figure 1. We note that the baking step (200 °C, overnight) in the oven was crucial for producing graphene grids with clean surfaces (Figure S2). The boiling point of the PMMA rinsing solvent (i.e., acetone) is 56 °C. However, we found that a moderate baking temperature (100 °C) was not sufficient to remove the swelled solvent in the PMMA residues (Figure S2a). We recommend performing the baking step without vacuum assistance as the nitrogen and oxygen species facilitate the removal of PMMA residues during this process (Figure S2b).53 By increasing the temperature to 200 °C, we minimized the residual PMMA on the graphene surface without damaging the coated graphene (Figure S2c).54 The extension of baking time to 48 h barely improved cleanness (Figure S2d). Although residual PMMA on the graphene surface is inevitable, because it adheres to surface defects/boundaries during graphene synthesis by CVD, it can be minimized by the high-temperature baking process.55 We also noticed that two layers of protection (the lid of the glass Petri dish and the beaker) during overnight baking prevent direct exposure to heat and thus minimize the damage to the graphene and holey carbon. Both copper and gold Quantifoil grids were examined to test which grid material is more suitable for the graphene coating procedure (Figure S3). We found that the Au Quantifoil grid exhibits better graphene coating because the Cu Quantifoil grid becomes heavily oxidized during baking, as indicated by color change and more damage to the graphene/Quantifoil holey carbon during overnight baking at 200 °C.
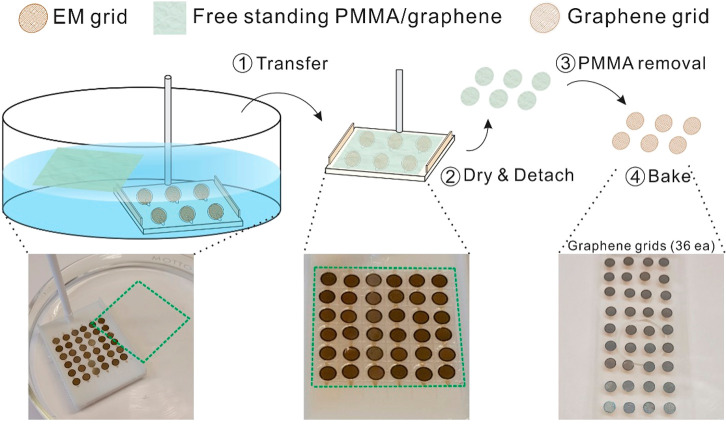
Figure 1.
Schematic view and photographs of graphene grid production. Green dashed squares in photographs denote the PMMA/graphene region. First, 36 Au Quantifoil grids were placed in the 3D-printed grid transfer tool (Figure S1) immersed in the water. The 2.5 cm × 2.5 cm PMMA-coated graphene layer is floated on the water and coated on the Au Quantifoil grid by lifting the grid transfer tool using the cylinder bar attached in the grid transfer tool (1, Transfer). Second, grids are dried using an oven (100 °C; preheated, 30 min) and individually detached/immersed in the acetone solvent (2, Dry & Detach; and 3, PMMA removal). Finally, 36 grids were placed on the cover slide and baked in an oven at 200 °C (preheated) overnight (4, Bake).
Characterization of the Graphene Grid
Previous studies in generating graphene-/graphene-oxide-coated grids have demonstrated the coating quality by testing and visualizing standard macromolecules such as the 30S ribosomal subunit and apo-ferritin using cryo-EM.25,35,36 Although this approach may indirectly indicate the quality of graphene or graphene-oxide grids, some or all of the cryo-EM data must be used to examine the quality of the grids, which is time-consuming and poses a potential risk to the precious specimen on the grid. Therefore, easily applied quality control and validation tools are needed before sample application. As quality control tools for graphene grids, we employed four approaches: SEM, bright-field transmission electron microscopy (BF-TEM), Raman spectroscopy, and AFM.
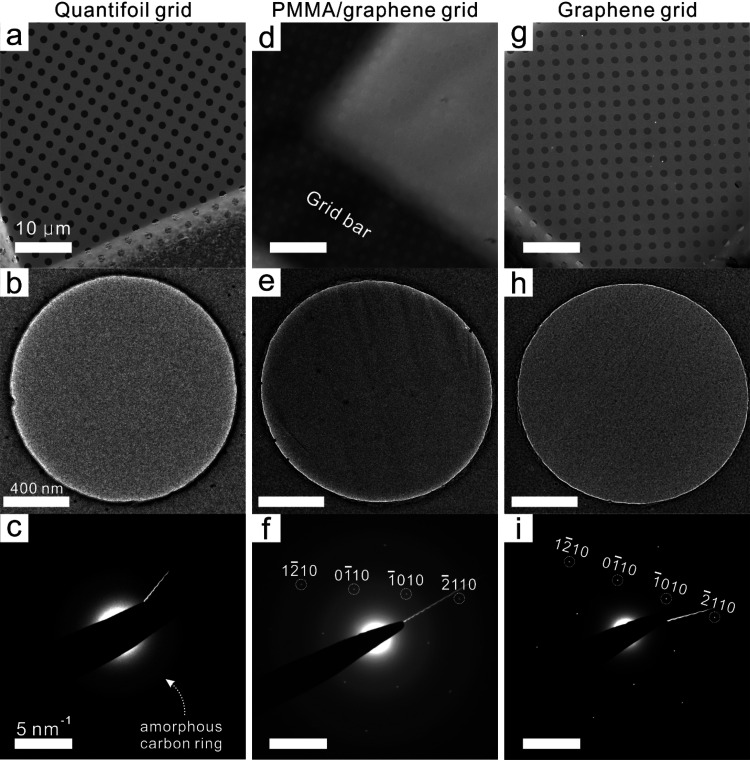
The graphene-coated Au Quantifoil grids were characterized by using SEM and BF-TEM to examine the coverage and surface quality. First, we compared the untreated PMMA/graphene-coated Au Quantifoil grids (before PMMA removal) and the PMMA-free graphene grids. As shown in Figure 2d, the surface of the PMMA/graphene grid was fully covered with a thick PMMA layer (approximately 500 nm according to the product information), which even blocked the observation of the hole patterns in the commercial Quantifoil grid (Au 300 mesh, R1.2/1.3). The TEM image and selected area electron diffraction (SAED) pattern of the PMMA/graphene grid (Figure 2e,f) showed a strong scattering pattern of amorphous carbon, which originated from the covered PMMA layer, and the characteristic SAED pattern of monolayer graphene (Figure 2f).56−58 After PMMA removal by rinsing and baking, most of the thick PMMA layer was eliminated from the graphene surface, as confirmed by SEM, BF-TEM, and SAED observations (Figure 2g–i). The SEM image indicates that the graphene fully covers the hole-patterned carbon of the Au Quantifoil grid. The SAED diffraction image further demonstrates that the diffraction pattern of the amorphous carbon ring is minimized, except for residual signals originating from the supporting holey carbon film of the Quantifoil grid. The graphene transfer method seems to be applicable to various other types of EM grids, but the Au Quantifoil showed the best graphene coverage (>95%) and cleanness. We note that materials with larger mesh number and smaller hole diameter showed better graphene coverage and less damage to the graphene grids during baking. Smaller hole diameters have also been shown to minimize the beam-induced particle motion.9 BF-TEM images and SAED patterns of samples in the rinsing steps were compared to further visualize the cleanness and validate the single-layered graphene on the Au Quantifoil grid (Figure S4). Selected area aperture (800 nm) was carefully positioned to only collect diffractions from the suspended graphene region without any amorphous carbon background after rinsing steps (Figure S4a–c). In the PMMA/graphene-coated grid, the SAED pattern displayed the bright background signal coming from the amorphous carbon of the thick PMMA layer (Figure S4d). After rinsing 3 times in acetone and a backing step, only graphene peak signals were detected (Figure S4e,f), which further supports the cleanness of the graphene surface after treatment. To confirm graphene is a monolayer, 3D electron diffraction was employed (Figure S5).59 The 3D electron diffraction probed reciprocal out-of-plane (kz) structure of Bragg rods and confirmed that the specimen is a monolayer with distinctive out-of-plane features (Figure S5b,c). The high-resolution TEM image showed that the graphene monolayer coexists with graphene oxides and some amorphous carbon generated by the rinsing and baking processes (Figure S6).
Figure 2.
SEM, TEM, and SAED images. (a–c) Left-side images are from untreated commercial Au Quantifoil grids. (d–f) Middle-column images are from PMMA/graphene-coated Au Quantifoil grids. (g–i) Right-side images are PMMA-free graphene grids after rinsing/baking steps. (a, d, g) Scale bars in SEM images represent 50 μm. (b, e, h) Scale bars in TEM images represent 400 nm. (c, f, i) Scale bars in SAED images represent 5 nm–1.
Figure 3 shows the Raman spectra of commercial monolayer graphene on Cu foil (Graphenea; top), the commercial Quantifoil grid (Electron Microscopy Sciences; middle); and the in-house-built graphene-coated grid (bottom). Single-layer graphene without defects was confirmed in the commercial monolayer graphene on Cu foil by observing the absence of the D peak (1348 cm–1; defect-induced second-order Raman scattering) as well as the high peak intensity ratio between the G peak (1580 cm–1; in-plane vibrations of sp2-hybridized carbon atoms) and the 2D peak (2670 cm–1; second-order overtone of a different in-plane vibration).60,61 It is generally acknowledged that a Raman intensity ratio of the 2D and G peaks (I2D/IG) of >2.0 denotes single-layer graphene.62,63 The Raman spectra of the commercial Quantifoil grid (before the graphene transfer) show large, broad D and G bands, which are derived from the hole-patterned amorphous carbon (62). After graphene transfer to the Quantifoil grid, the I2D/IG was 7.2, which coincides with other characterizations of single-layered graphene (Table S1). Raman spectra of the graphene layer and the D and G band patterns of the Quantifoil grid appear in the graphene-coated grid, indicating that the graphene monolayer was successfully transferred and coated on the Quantifoil grid.
Figure 3.
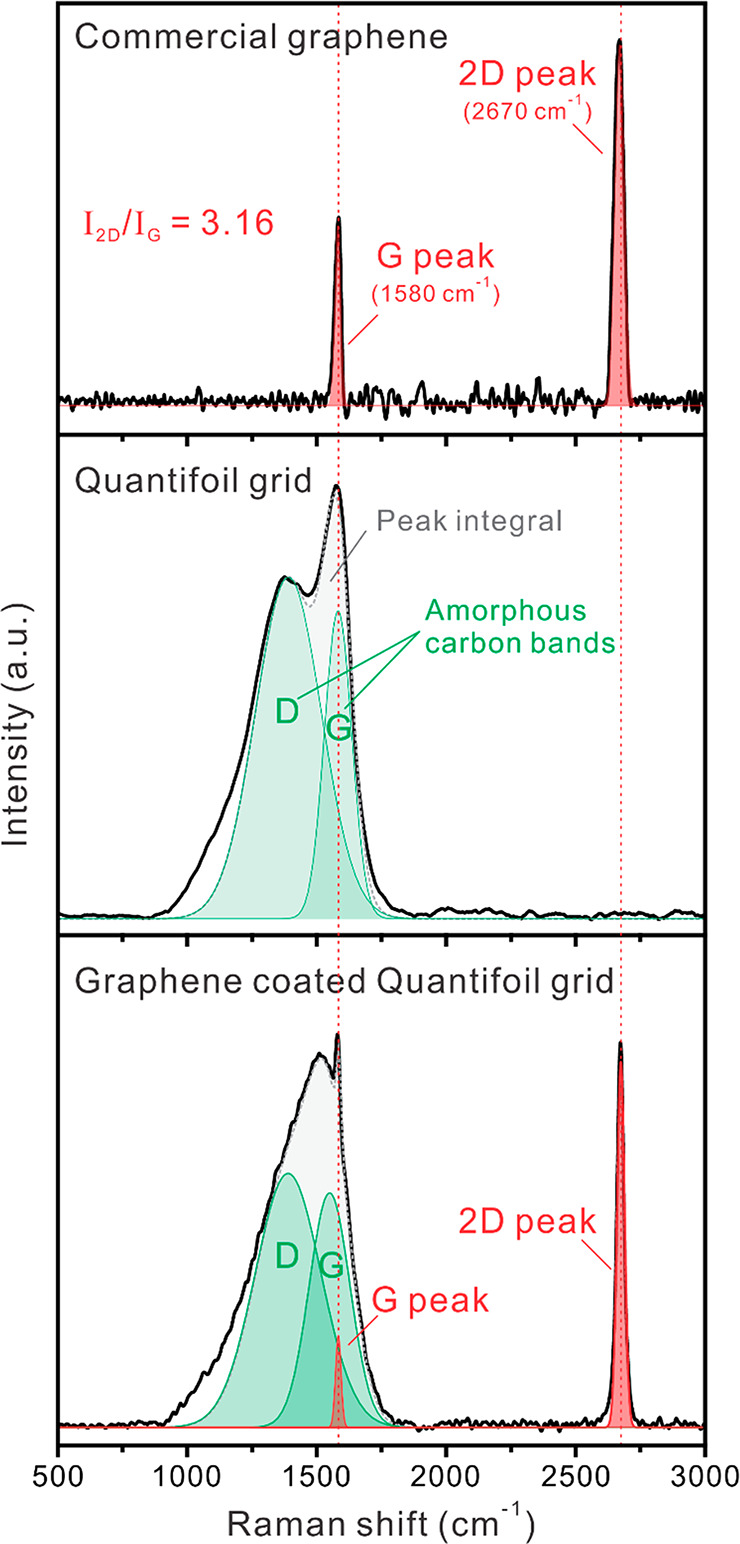
Raman spectra of the commercial graphene monolayer (top), commercial Au Quantifoil grid (middle), and graphene-coated Au Quantifoil grid (bottom). The red-dotted vertical line denotes the peak positions (G and 2D) of graphene. Green regions denote the Raman band positions (D and G) of amorphous carbon.
AFM was further employed to characterize the morphology and physical properties of the graphene grids. The Quantifoil grid has regularly patterned holes that were fully covered after the PMMA/graphene transfer process (Figure 4a,c). After the thick PMMA layer was removed, the hole pattern covered by graphene appeared, with minimal surface residues (Figure 4e,f); most of the areas of interest (cryosample positions) were clean and defect-free. The graphene grids exhibited a relatively large z-scale variation (300 nm), mainly because of a fragile grid wrinkle that occurred during AFM sample preparation (taping and flattening on the glass substrate). Inside of the graphene-covered holes, the surface was flat and clean and showed low z-scale variation (10 nm) (Figure 4g,h). Therefore, the Raman spectroscopy and AFM results, in addition to SEM and TEM images, can be good validation tools for examining the quality of graphene-coated grids after graphene transfer and before specimen application.
Figure 4.
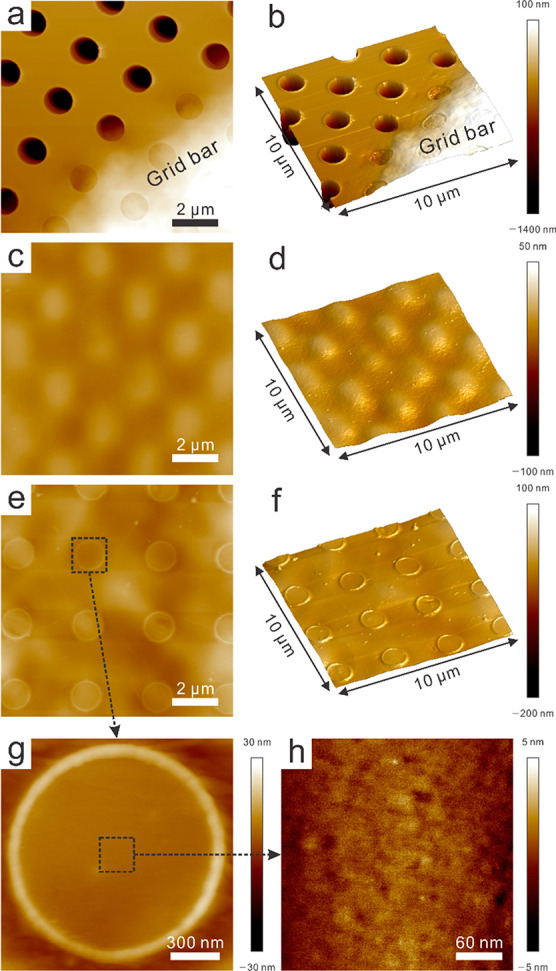
AFM height mode and 3D mode images of the Quantifoil, PMMA/graphene-coated Au Quantifoil, and PMMA-free graphene grids. Note that the z-scale is not the same in these images. (a) Height mode and (b) 3D image of Quantifoil grid with 1.5 μm z-scale. (c) Height mode and (d) 3D image of PMMA/graphene-coated Quantifoil with 150 nm z-scale. (e) Height mode and (f) 3D image of PMMA-free graphene grid with 300 nm z-scale. Magnified region of the black dotted box for the (g) single hole and (h) center of the hole with 60 and 10 nm z-scales, respectively.
Cryo-EM Structure Determination of M. capsulatus sMMOH
Since Rosenzweig and co-workers determined the first crystal structure of M. caps sMMOH in 1993, the structures of different enzymatic stages of sMMOH have been determined by X-ray crystallography.41,64−66 Earlier studies indicated that the geometry of the nonheme diiron center plays a critical role in indicating the enzymatic stage of sMMOH. The diiron geometry, which is coordinated by two histidines (His147 and His246) and four glutamates (Glu114, Glu144, Glu209, and Glu243), undergoes subtle yet significant conformational changes depending on its catalytic reaction stage and its association with auxiliary subunits (e.g., MMOB and MMOD).44,46 This rearrangement of the diiron coordination is interlocked and triggered by the movement of the four-helix bundle (helices B, C, E, and F) that contain these two histidines and four glutamates. Although currently available crystal structures of sMMOH have provided valuable information regarding diiron coordination shifts under various circumstances, protein crystallization requires high precipitant concentrations and unnatural compounds, and the protein structure can be further constrained by crystal contacts. To illustrate the structure of sMMOH in solution and validate the general applicability of the graphene-coated grid, we determined the cryo-EM structures of M. caps sMMOH at resolutions of 2.9 and 2.5 Å using the Au Quantifoil and graphene-coated grids, respectively, under normal buffer conditions [30 mM HEPES (4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid), pH 7.0, 150 mM NaCl, and 1 mM TCEP (tris(2-carboxylethyl)phosphine)] (Figure 5a–c and Figure S7).
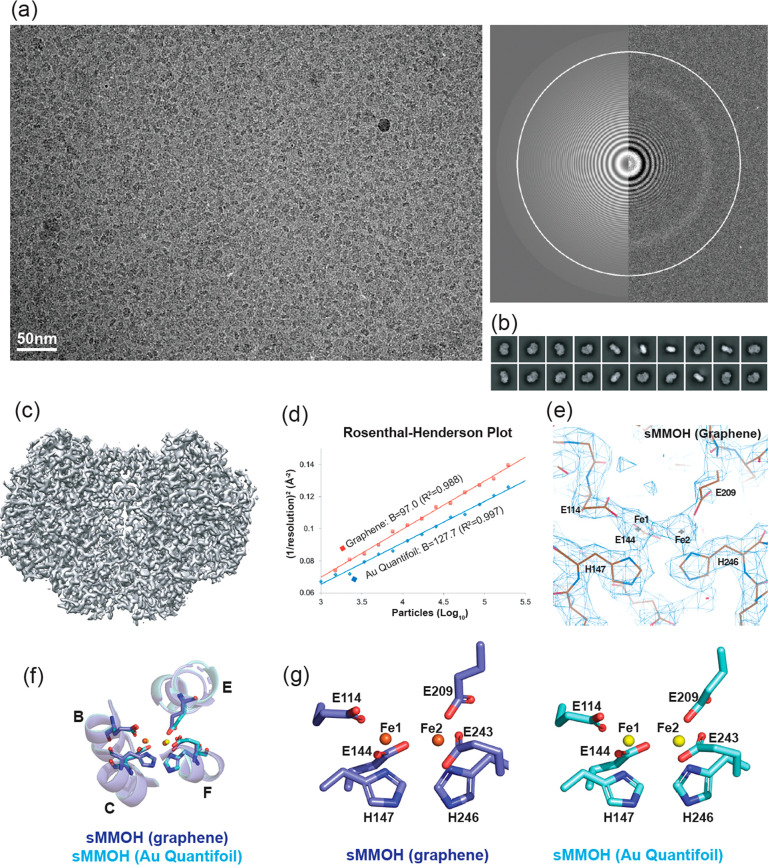
Figure 5.
(a) Representative microscopic image of M. caps sMMOH in the graphene grid (300 kV) with the image of the power spectrum. (b) Top 20 2D classes selected from 200 classes of sMMOH in the graphene grid. (c) Reconstructed 3D cryo-EM map (2.5 Å resolution) of sMMOH in the graphene grid. (d) Rosenthal–Henderson plot of sMMOH structures determined using the Au Quantifoil (blue diamond-shaped) and graphene (red square) grids. Dashed lines indicate the particles used for the final refinement. (e) Cryo-EM map of the nonheme diiron center of sMMOH (graphene grid) coordinated by four glutamates and two histidines. The image was captured by COOT. (f) Structural overlay of sMMOH four-helix bundle (helix B, C, E, and F) and (g) coordinating residues (four glutamates and two histidines) determined by Au Quantifoil (cyan; yellow for two irons) and graphene (blue; orange for two irons) grids.
Structural Comparison of sMMOH Determined Using Au Quantifoil and Graphene-Coated Grids
Purified sMMOH at concentrations of 1.3 and 0.5 mg/mL was applied to the Au Quantifoil and graphene-coated grids, respectively (Table S2). A total of 3075 (Au Quantifoil grid) and 3880 (graphene grid) raw movie stacks were collected using 300 kV Titan Krios with a K3 direct electron detector equipped with a BioQuantum energy filter (Figure 5a and Figure S7a). Particles of 2.1 M (Au Quantifoil grid) and 4.2 M (graphene grid) were obtained after particle selection by using the TOPAZ program.67 After iterative 2D and 3D classification using cryoSPARC (Figure 5b and Figure S7b), 843k (20.0% of total particles) and 389k (18.5% of total particles) particles were selected for the Au Quantifoil grid and graphene grid to reconstitute the cryo-EM structures of sMMOH at resolutions of 2.9 and 2.5 Å, respectively, after real-space refinement by PHENIX (Table S3, Figure 5c, and Figure S7c).59,68 The 2.5 Å cryo-EM structure of sMMOH obtained using the graphene grid shows clear side chain densities throughout the entire structure, including the nonheme diiron center (Figure 5e). The Rosenthal–Henderson plot indicates that the quality and behavior of the final selected particles from the graphene grid are slightly better than those of the particles from the Au Quantifoil grid (Figure 5d). We also noted that the graphene grid requires approximately five times less protein to obtain a similar number of particles (Table S2). Although we found that sMMOH exhibited a preferred orientation on the graphene grid, this orientation might be caused in part by the molecular nature of sMMOH, which has a flat shape (Figure 5b,c and Figure S8). It is not clear why the specimen on the graphene grid exhibits better behavior than that on the Au Quantifoil grid, but one possible explanation might be the minimal particle exposure to the AWI. Because particles adhere to the graphene surface after plasma treatment, exposure to the AWI can be dramatically reduced. Russo and Passmore also demonstrated that a plasma-treated graphene grid reduces the beam-induced particle motion, which may further contribute to resolution improvement.31 However, the final refined structures of sMMOH on the Au Quantifoil and graphene grids are almost identical, suggesting that structural perturbation by the graphene layer is negligible (Figure S7d). The Cα root-mean-square (rms) difference in the two structures is 0.338 Å, and they have nearly identical diiron center geometries (Figure 5f,g and Figure S7d).
Particle Distribution of sMMOH on Graphene-Coated Grids Using Cryoelectron Tomography
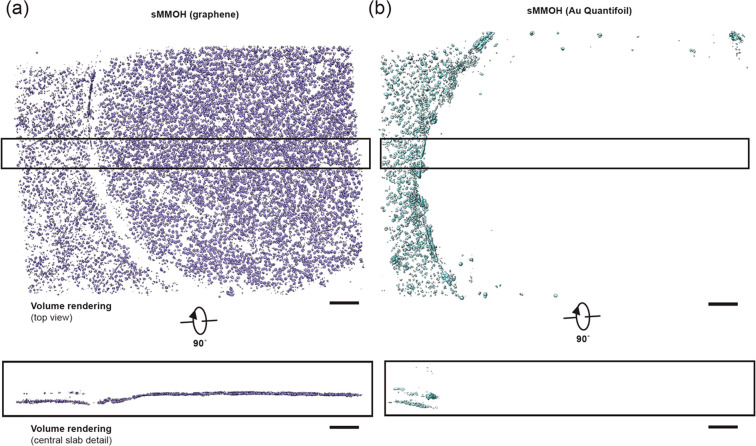
Particles tend to adhere to the oxidized hydrocarbon surface after plasma treatment of graphene or an amorphous carbon layer on girds. This allows the particles to adsorb to the grid surface during cryo-EM and negative-stain EM sample preparation. Since particles are not exposed to the air–water interface after attachment to the oxidized graphene layer, this further provides an additional advantage in data quality by preventing protein denaturation induced by the air–water interface. To visualize the distribution of particles on graphene-coated grids, we collected a tilt series of plunge-frozen sMMOH particles on graphene-coated and Au Quantifoil grids, respectively. As shown in Figure 6, sMMOH particles are densely populated on the graphene surface. The same protein concentration of sMMOH (0.24 mg/mL) was applied to prepare the Au Quantifoil grid, but most of the particles were attracted to the amorphous carbon and barely detectable within the grid hole (Figure S9, Movies S2 and S3). This observation agrees well with the previous cryogenic-electron tomography (cryo-ET) result of 1-pyrene carboxylic acid on chemically oxidized graphene-coated grids.69 Therefore, the cryo-ET data of the graphene-coated grid further support that the graphene-coated grid not only concentrates particles on the surface but also prevents the protein denaturation mediated by the air–water interface.
Figure 6.
M. caps sMMOH particle distribution on plunge-frozen graphene-coated and Au Quantifoil grids. (a) Top and central slab views of segmented sMMOH particles (purple) from a tomogram of a graphene-coated grid. (b) Top and central slab views of segmented sMMOH particles (cyan) from a tomogram of a Au Quantifoil grid. Scale bar is 100 nm.
Structural Comparison of sMMOH Determined by Cryo-EM and X-rays
Purified sMMOH was crystallized under buffer conditions of 25 mM Li2SO4, 50 mM NH4OAc, and 5% polyethylene glycol (PEG 4000), and its structure was determined at a resolution of 1.7 Å. The structural overlay of the crystal structure (PDB ID: 1MTY) and cryo-EM structure (graphene grid) of sMMOH exhibits a Cα rms difference of 0.351 Å, indicating that the overall architecture of the two structures is very similar (Figure S10). However, we observed differences in the diiron position and surrounding diiron coordinating residues (Figure 7). First, we observed differences in the positions of the two irons. Fe13+ and Fe23+ were shifted by 0.5 Å (protomer1)/0.2 Å (protomer2) and 1.1 Å/0.6 Å, respectively, in the cryo-EM structure compared to the X-ray structure (Figure 7). The distance between the two γ carboxyl groups of Glu114 and Glu243 also changed, from 5.5 Å/4.4 Å (protomer1) and 5.6 Å/4.6 Å (protomer2) in the X-ray structure to 5.5 Å/5.5 Å (protomer1) and 5.9 Å/5.4 Å (protomer2) in the cryo-EM structure. This result indicates that two γ carboxyl groups of Glu114 and Glu243 moved away from each other in the cryo-EM structure, which resulted in changes in the positions of the two irons, particularly that of Fe2. The iron–iron distances also increased from 3.0 Å (protomer1) and 3.1 Å (protomer2) to 3.2 and 3.2 Å, respectively. The difference we observed here can be derived from the crystal packing since the geometry of the diiron center is greatly influenced by the four-helix bundle (Figure 5f; helices B, C, E, and F), whose conformation might be restricted by the crystal packing in the X-ray structure. We further observed an unidentified cryo-EM density on top of the diiron center, where the substrates had been located (Figure S11).46,59,66 A similar density was also observed in several crystal structures of sMMOH, but one of the crystallization compounds was assigned to refine the structure (e.g., acetate, in PDB ID: 1MMO).41 This result indicates that this increase in density might be attributable to one of the sMMOH substrates or molecules involved in the catalytic reaction of sMMOH, which might be copurified during protein purification. Regardless of the type of compound, it certainly participates in diiron coordination in the resting state of sMMOH (Figure S11). Because we were not able to identify the nature of this compound, we decided not to build a model of this density.
Figure 7.
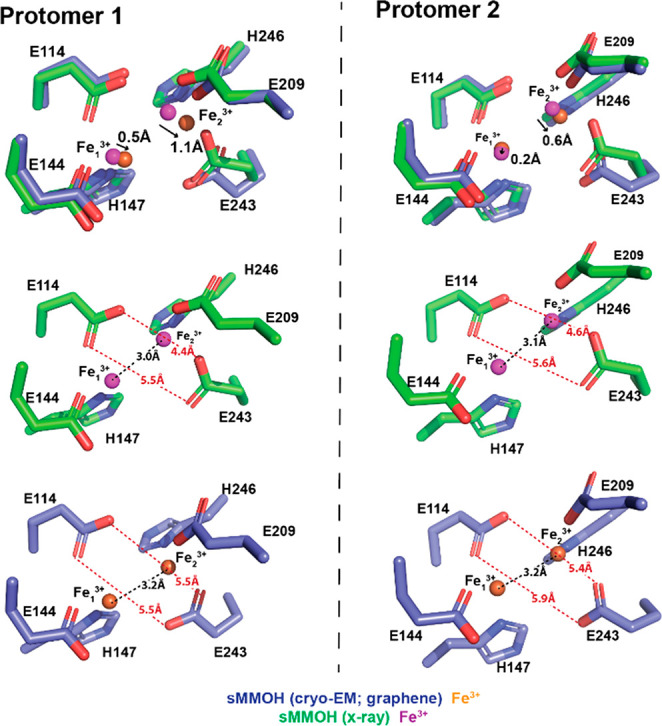
Nonheme diiron center geometry comparison between the X-ray structure (PDB ID: 1MTY) and cryo-EM structure (graphene grid). The diiron center geometry of both sMMOH protomers (protomers 1 and 2) was compared.
Conclusions
In this paper, a versatile method of developing graphene-coated EM grids (i.e., graphene grids) was established using various in situ characterization tools to assess the quality. This method does not require professional skills or expensive instruments and can be easily applied to commercially available EM grids with minimal polymeric residues and high coverage. Moreover, high-quality graphene coatings were obtained after cleaning and baking steps. The graphene grids were tested by determining the cryo-EM structure of M. caps sMMOH at a resolution of 2.5 Å. Therefore, we illustrate that our method ensures both the quality and quantity of the graphene grids (with a capacity of 36 at once), making it convenient to produce.
Experimental Methods
Protocol for Graphene Transfer to Cryo-EM Grid
A 25 × 25 mm poly(methyl methacrylate) (PMMA)/graphene pad (Trivial Transfer Graphene, SKU# TTG10011) was purchased from ACS Material and stored in a 2–8 °C refrigerator before use. The graphene transfer process was based on user instructions with several modifications. First, 36 gold Quantifoil holey carbon grids (Au R1.2/1.3 Quantifoil grids; the amorphous carbon layer facing up) were placed on a three-dimensional (3D)-printed grid transfer tool (Figure S1) immersed in deionized water in a Petri dish, and the PMMA/graphene/support textile pad was gently placed on the water surface to prevent water overflow onto the graphene layer. Direct water overflow onto the graphene layer will cause it to shrink, making it unusable. The supporting textile was gently pushed to the bottom of the Petri dish using tweezers. This procedure separated the graphene layer from the textile pad and caused the graphene layer to float on the water surface (Figure 1, Movie S1). Then the PMMA/graphene layer was carefully matched with the top of the 36 grids using tweezers, and the structure was slowly removed from the water by lifting the grid transfer tool. Residual water in the grid transfer tool was drained with a cleaning wipe, and the PMMA/graphene/grid structure was placed in an oven (preheated; 100 °C, 30 min) to dry completely. This step also increased the contact between the graphene layer and the grids. Fully dried PMMA/graphene-coated grids were individually detached from the grid transfer tool and immersed in acetone solvent (50 °C, 30 min, repeated three times with mild stirring) in a Petri dish to dissolve and eliminate the PMMA layer (do not use any plastic/polymer materials to touch the acetone solution). The acetone-free, air-dried, PMMA-free graphene grids were transferred to a microscope slide glass and baked in the oven (preheated; 200 °C, 24 h) to evaporate the remaining solvent and swell the polymers on the graphene. To prevent direct exposure of the graphene grids to heat, we placed the slide glass in a Petri dish (glass, with a lid) and also covered it with a glass beaker during baking. The generated graphene grids were individually transferred to a grid box, which was held in a desiccator for up to several months before use. All procedures described here are also available in Movie S1 and the following link (http://www.uscho-lab.com/research/).
Characterizations of Graphene Grids
Scanning electron microscopy (SEM) images were obtained by using a NOVA NanoLab 200 instrument with an electron beam power of 5 kV at 0.40 nA. The graphene and other grids were placed on a grid-holding accessory for SEM observation. Transmission electron microscopy (TEM) images were obtained at 200 kV by using a JEOL-2010F instrument. Raman spectra were collected using a Renishaw instrument equipped with a 532 nm diode laser having a power of 0.5 W; a 1200 lines/mm grating and an Olympus SLMPlan 20× objective lens were also used. All spectra were obtained in the Raman shift range of 1000–3000 cm–1 to analyze the framework bands and at peak positions of 2680 cm–1 to analyze the 2D band and 1580 and 1380 cm–1 to monitor the G and D peaks of graphene (amorphous carbon bands), respectively. The laser was calibrated in static scan mode by using a silicon standard.
TEM, SAED, and 3D Electron Diffraction
S/TEM and SAED were performed on a Thermo Fisher Talos instrument operated at 200 kV equipped with a Gatan OneView camera. For 3D electron diffraction, 31 SAED patterns were collected across a −15° to +15° range and then mapped to 3D reciprocal space to reveal that the graphene was monolayer.
Atomic Force Microscopy (AFM)
AFM images were taken using a Veeco Dimension Icon atomic force microscope with a ScanAsyst-Air AFM tip from Bruker Nano Inc. The sample grids were held on a slide glass by copper tape/slide glass during scanning with a vacuum-assisted lock on slide glass. The data were analyzed by using Nanoscope Analysis 2.0 software. The AFM tip was calibrated in thermal tuning mode with a spring constant of 0.4 N/m.
Purification of M. capsulatus sMMOH
M. caps (Bath) sMMOH was purified as described in the literature.44 In brief, M. caps (Bath) was cultured in a nitrate mineral salt medium at 30 °C. Then the harvested cells were collected by centrifugation (11,300g) for 20 min at 4 °C. The cell pellet suspended in the buffer was lysed at 4 °C (CV334, Sonics), and the lysate was centrifuged at 30,000g for 45 min at 4 °C. The supernatant was collected and filtered through a 0.22 μm membrane. The filtrate was loaded onto DEAE Sepharose, Superdex 200, and finally Q Sepharose columns to obtain sMMOH with >95% purity.
Cryo-EM Sample Preparation on Graphene Grids
Before sample loading, the graphene grids were treated by glow discharge (PELCO easiGlow, Ted Pella Inc.) at 5 mA for 60 s in vacuum (<0.26 mbar). Cryo-EM sample grids were prepared in a Vitrobot Mark IV instrument (Thermo Fisher Scientific), which was set to 4 °C at 100% humidity. Purified protein solution (3 μL) was loaded onto the graphene surface of the graphene grid or holey carbon surface of the Au Quantifoil grids as indicated. After 30 s, the grids were blotted for 4 s with a blot force of 0 and immediately plunged into precooled liquid ethane for vitrification.
Data Acquisition
A total of 3880 (graphene grid) and 3075 (Au Quantifoil grid) raw movie stacks were automatically collected on a 300 kV Titan Krios using a K3 direct electron detector (with a BioQuantum energy filter, Gatan) at the Pacific Northwest Center for Cryo-EM (PNCC). Raw movies from Quantifoil and graphene grid were collected in K3 super-resolution mode at a magnification of 81,000× (slit width 20 eV, spot size 5, C2 aperture 50 μm) with a super-resolution pixel size of 1.027 and 1.059 Å, respectively. The total exposure time was 3.4 s at 0.85 e–/Å2 per frame to generate 60-frame gain-normalized microbial reverse-electrodialysis cell stacks. The total dose per stack was 49 and 51 e–/Å2 for Quantifoil and graphene grid, respectively.
Cryo-EM Data Processing and Model Refinement
All processing was completed in RELION and cryoSPARC.2,70 The MotionCor2 implemented in RELION corrected the initial drift and beam-induced motion, and Gctf was used to measure the contrast transfer function (CTF).71 Following CTF estimation, micrographs were manually inspected, and those with outliers in the defocus value, ice thickness, or astigmatism, as well as micrographs with lower predicted CTF-correlated resolution (>5 Å), were removed from further processing. The initial set of particles was selected by using Topaz.67 The selected particles were further 2D- and 3D-classified by iterative classification and selection rounds using an ab initio 3D reconstructed model as a starting reference in cryoSPARC. A total of 843k and 389k particles were chosen to build final 3D reconstruction maps with a resolution of 2.32 and 2.75 Å (FSC 0.143) for the graphene grid and Au Quantifoil grid, respectively (Figure S12). Using the crystal structure of sMMOH (PDB ID: 1MTY), we refined the structures using real-space refinement in the PHENIX program and COOT.68,72,73 The reported resolutions of the final maps (2.5 Å for the graphene grid and 2.9 Å for the Au Quantifoil grid) were estimated using the map versus model FSC curves (FSC0.5map-vs-model) in phenix.xtriage.68 UCSF Chimera was used to visualize the EM density and obtain illustrations for figures.74
Cryoelectron Tomography
Both Au Quantifoil and graphene-coated grids were prepared with the same concentration of sMMOH (0.24 mg/mL). Vitrified specimens were imaged with a 300 kV Titan Krios G4i transmission electron microscope equipped with a K3 summit direct electron detector and a Gatan energy filter. The magnification was set to a pixel size of 2.122 Å for the tilt series data collection. Dose-fractionated images were automatically recorded using SerialEM in counting mode from −60° to +60° using a dose-symmetric acquisition scheme with a total dose of 82 e–/Å2.75,76 The tilt series was processed with IMOD.77 Tilt series were aligned by patch tracking, and tomograms were reconstructed by weighted back projection as implemented in IMOD. Tomograms were segmented with the convolutional neural network method implemented in EMAN2.2.78 UCSF Chimera was used for visualizing the tomographic volumes.79 Software used for processing tilt series and visualizing volumes was curated by SBGrid.80
Acknowledgments
We thank Dr. Leila Foroughi and Dr. Adam Matzeger in the Department of Chemistry at the University of Michigan for the use of the Raman spectroscopy instrument and assistance. We thank the staff at the University of Michigan cryo-EM center for the use of the Morgagni instrument and assistance. We thank Dr. Haiping Sun and the Michigan Center for Materials Characterization for the use of the AFM instrument and assistance. Dr. Seung-Jae Lee at the Chonbuk National University kindly provided the purified M. caps sMMOH.
Glossary
Abbreviations
- cryo-EM
cryogenic electron microscopy
- M. caps
Methylococcus capsulatus
- sMMOH
soluble methane monooxygenase hydroxylase
- AWI
air–water interface
- 2D
two-dimensional
- CVD
chemical vapor deposition
- pMMO
particulate methane monooxygenase
- sMMO
soluble methane monooxygenase
- sMMOR
sMMO reductase
- sMMOB
sMMO regulatory subunit
- sMMOD
sMMO inhibitory subunit
- NADH
nicotinamide adenine dinucleotide
- BF-TEM
bright-field transmission electron microscopy
- SAED
selected area electron diffraction
- rms
root-mean-square
- cryo-ET
cryogenic electron tomography
- SEM
scanning electron microscopy
- AFM
atomic force microscopy
- CTF
contrast transfer function
- FSC
Fourier shell correlation
- PMMA
poly(methyl methacrylate)
Supporting Information Available
. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.3c00463.
Figures showing additional 3D-printed grid transfer tool, optical microscopy and SEM images, BF-TEM images and SAED patterns, Raman spectroscopy results, AFM results, 2D classification results, particle orientation and distribution images, cryo-EM density map of sMMOH, and FSC curves; and tables for Raman deconvolution results, statistics for cryo-EM data collection, refinement, and validation (PDF)
Movie S1: Batch-mode graphene grid production video (MOV)
Movie S2: Tomogram of sMMOH particles on a graphene-coated grid (MOV)
Movie S3: Tomogram of sMMOH particles on a Au Quantifoil grid (MOV)
Author Contributions
∇ E.A. and B.K. contributed equally. The manuscript was written through contributions of all authors. U.-S.C. designed the research. E.A. and B.K. performed research and analyzed data. S.P. performed experiments in revision. A.L.E. and S.M. performed the cryoelectron tomography data collection and analysis. S.H.S. and R.H. performed the TEM and SAED measurements. E.A., B.K., and U.-S.C. wrote the manuscript. All authors have given approval to the final version of the manuscript.
This work was supported by grants from R01 DK111465, R01 CA250329, and R01 NS116008 to U.-S.C. A portion of this research was supported by NIH grant U24GM129547, performed at the PNCC at OHSU, and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
Pre-print submission: Ahn, E.; Kim, B.; Park, S.; Erwin, A. L.; Sung, S.H.; Hovden, R.; Mosalaganti, S.; Cho, U.S. Batch Production of High-Quality Graphene Grids for Cryo-EM: Cryo-EM Structure of Methylococcus capsulatus Soluble Methane Monooxygenase Hydroxylase. bioRxiv, 2021.12.26.474209, 2022. DOI: 10.1101/2021.12.26.474209 (accessed July 10, 2022).
The authors declare no competing financial interest.
Supplementary Material
References
- Dubochet J. Cryo-EM-The First Thirty Years. J. Microsc 2012, 245, 221–224. 10.1111/j.1365-2818.2011.03569.x. [DOI] [PubMed] [Google Scholar]
- Scheres S. H. RELION: Implementation of a Bayesian Approach to Cryo-EM Structure Determination. J. Struct. Biol. 2012, 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Cryo-EM Enters a New Era. Elife 2014, 3, e03678 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X. C.; McMullan G.; Scheres S. H. W. How Cryo-EM is Revolutionizing Structural Biology. Trends Biochem. Sci. 2015, 40, 49–57. 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Zivanov J.; Nakane T.; Forsberg B. O.; Kimanius D.; Hagen W. J.; Lindahl E.; Scheres S. H. New Tools for Automated High-Resolution Cryo-EM Structure Determination in RELION-3. Elife 2018, 7, e42166 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C. L.; Berman H. M.; Chiu W. Evolving Data Standards for Cryo-EM Structures. Struct. Dyn. 2020, 7, 014701. 10.1063/1.5138589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. J.; Cheng A.; Noble A. J.; Eng E. T.; Kim L. Y.; Carragher B.; Potter C. S. Routine Determination of Ice Thickness for Cryo-EM Grids. J. Struct. Biol. 2018, 204, 38–44. 10.1016/j.jsb.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner A. T.; Naydenova K.; Castro-Hartmann P.; Nguyen-Phan T. C.; Russo C. J.; Sader K.; Hunter C. N.; Cogdell R. J.; Qian P. The 2.4 A cryo-EM Structure of a Heptameric Light-Harvesting 2 Complex Reveals Two Carotenoid Energy Transfer Pathways. Sci. Adv. 2021, 7, eabe4650 10.1126/sciadv.abe4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenova K.; Jia P.; Russo Christopher J. Cryo-EM with Sub-1 Å Specimen Movement. Science 2020, 370, 223–226. 10.1126/science.abb7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser R. M.Specimen Behavior in the Electron Beam. In Methods in Enzymology; Crowther R. A., Ed.; Academic Press: Cambridge, 2016; Vol. 579, Chapter 2, pp 19–50. [DOI] [PubMed] [Google Scholar]
- Tan Y. Z.; Baldwin P. R.; Davis J. H.; Williamson J. R.; Potter C. S.; Carragher B.; Lyumkis D. Addressing Preferred Specimen Orientation in Single-Particle Cryo-EM Through Tilting. Nat. Methods 2017, 14, 793–796. 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. Orienting the Future of Bio-Macromolecular Electron Microscopy. Chinese Physics B 2018, 27, 063601. 10.1088/1674-1056/27/6/063601. [DOI] [Google Scholar]
- Drulyte I.; Johnson R. M.; Hesketh E. L.; Hurdiss D. L.; Scarff C. A.; Porav S. A.; Ranson N. A.; Muench S. P.; Thompson R. F. Approaches to Altering Particle Distributions in Cryo-Electron Microscopy Sample Preparation. Acta Crystallogr. D Struct. Biol. 2018, 74, 560–571. 10.1107/S2059798318006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Imprima E.; Floris D.; Joppe M.; Sanchez R.; Grininger M.; Kuhlbrandt W. Protein Denaturation at the Air-Water Interface and How to Prevent it. Elife 2019, 8, e42747 10.7554/eLife.42747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsiccio A.; McCarty J.; Pisano R.; Shea J. E. Heightened Cold-Denaturation of Proteins at the Ice-Water Interface. J. Am. Chem. Soc. 2020, 142, 5722–5730. 10.1021/jacs.9b13454. [DOI] [PubMed] [Google Scholar]
- Rhinow D.; Kühlbrandt W. Electron Cryo-Microscopy of Biological Specimens on Conductive Titanium-Silicon Metal Glass Films. Ultramicroscopy 2008, 108, 698–705. 10.1016/j.ultramic.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Rhinow D.; Buenfeld M.; Weber N. E.; Beyer A.; Golzhauser A.; Kuhlbrandt W.; Hampp N.; Turchanin A. Energy-Filtered Transmission Electron Microscopy of Biological Samples on Highly Transparent Carbon Nanomembranes. Ultramicroscopy 2011, 111, 342–349. 10.1016/j.ultramic.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Llaguno M. C.; Xu H.; Shi L.; Huang N.; Zhang H.; Liu Q.; Jiang Q. X. Chemically Functionalized Carbon Films for Single Molecule Imaging. J. Struct. Biol. 2014, 185, 405–417. 10.1016/j.jsb.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo C. J.; Passmore L. A. Progress Towards an Optimal Specimen Support for Electron Cryomicroscopy. Curr. Opin. Struct. Biol. 2016, 37, 81–89. 10.1016/j.sbi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Zhang L.; Wen Z.; Chen H.; Li S.; Ji G.; Yin C. C.; Sun F. Amorphous Nickel Titanium Alloy Film: A New Choice for Cryo Electron Microscopy Sample Preparation. Prog. Biophys. Mol. Biol. 2020, 156, 3–13. 10.1016/j.pbiomolbio.2020.07.009. [DOI] [PubMed] [Google Scholar]
- Kelly D. F.; Dukovski D.; Walz T. Monolayer Purification: a Rapid Method for Isolating Protein Complexes for Single-Particle Electron Microscopy. Proc. Natl. Acad. Sci. U S A 2008, 105, 4703–4708. 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. F.; Dukovski D.; Walz T. Strategy for the Use of Affinity Grids to Prepare Non-His-Tagged Macromolecular Complexes for Single-Particle Electron Microscopy. J. Mol. Biol. 2010, 400, 675–681. 10.1016/j.jmb.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. G.; Walton R. W.; Song A.; Hwu P.; Stubbs M. T.; Yannone S. M.; Arbelaez P.; Dong M.; Glaeser R. M. Electron Microscopy of Biotinylated Protein Complexes Bound to Streptavidin Monolayer Crystals. J. Struct. Biol. 2012, 180, 249–253. 10.1016/j.jsb.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Yu G.; Li K.; Huang P.; Jiang X.; Jiang W. Antibody-Based Affinity Cryoelectron Microscopy at 2.6-A Resolution. Structure 2016, 24, 1984–1990. 10.1016/j.str.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin C. J.; Wright K. J.; Bolton S. C.; Hyun S. H.; Krynski K.; Grover M.; Yu G.; Guo F.; Kinzer-Ursem T. L.; Jiang W.; Thompson D. H. Selective Capture of Histidine-tagged Proteins from Cell Lysates Using TEM grids Modified with NTA-Graphene Oxide. Sci. Rep. 2016, 6, 32500. 10.1038/srep32500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov K. S.; Geim A. K.; Morozov S. V.; Jiang D.; Zhang Y.; Dubonos S. V.; Grigorieva I. V.; Firsov A. A. Electric Field Effect in Atomically Thin Carbon Films. Science 2004, 306, 666–669. 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- Lee C.; Wei X.; Kysar J. W.; Hone J. Measurement of the Elastic Properties and Intrinsic Strength of Monolayer Graphene. Science 2008, 321, 385–388. 10.1126/science.1157996. [DOI] [PubMed] [Google Scholar]
- Bai J.; Zhong X.; Jiang S.; Huang Y.; Duan X. Graphene Nanomesh. Nat. Nanotechnol. 2010, 5, 190–194. 10.1038/nnano.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko A. N.; Polini M.; Novoselov K. S. Graphene Plasmonics. Nat. Photonics 2012, 6, 749–758. 10.1038/nphoton.2012.262. [DOI] [Google Scholar]
- Lee G. H.; Cooper R. C.; An S. J.; Lee S.; van der Zande A.; Petrone N.; Hammerberg A. G.; Lee C.; Crawford B.; Oliver W.; Kysar J. W.; Hone J. High-Strength Chemical-Vapor-Deposited Graphene and Grain Boundaries. Science 2013, 340, 1073–1076. 10.1126/science.1235126. [DOI] [PubMed] [Google Scholar]
- Russo C. J.; Passmore L. A. Controlling Protein Adsorption on Graphene for Cryo-EM Using Low-Energy Hydrogen Plasmas. Nat. Methods. 2014, 11, 649–652. 10.1038/nmeth.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovcak E.; Wang F.; Zheng S. Q.; Yu Z.; Li S.; Betegon M.; Bulkley D.; Agard D. A.; Cheng Y. A Simple and Robust Procedure for Preparing Graphene-Oxide Cryo-EM Grids. J. Struct. Biol. 2018, 204, 80–84. 10.1016/j.jsb.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N.; Zhang J.; Chen Y.; Liu C.; Zhang X.; Xu K.; Wen J.; Luo Z.; Chen S.; Gao P.; Jia K.; Liu Z.; Peng H.; Wang H. W. Bioactive Functionalized Monolayer Graphene for High-Resolution Cryo-Electron Microscopy. J. Am. Chem. Soc. 2019, 141, 4016–4025. 10.1021/jacs.8b13038. [DOI] [PubMed] [Google Scholar]
- Fan X.; Wang J.; Zhang X.; Yang Z.; Zhang J. C.; Zhao L.; Peng H. L.; Lei J.; Wang H. W. Single Particle Cryo-EM Reconstruction of 52 kDa Streptavidin at 3.2 Angstrom Resolution. Nat. Commun. 2019, 10, 2386. 10.1038/s41467-019-10368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenova K.; Peet M. J.; Russo C. J. Multifunctional Graphene Supports for Electron Cryomicroscopy. Proc. Natl. Acad. Sci. U S A 2019, 116, 11718–11724. 10.1073/pnas.1904766116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Fan X.; Wang H.; Zhao F.; Tully C. G.; Kong J.; Yao N.; Yan N. High-Yield Monolayer Graphene Grids for Near-Atomic Resolution Cryoelectron Microscopy. Proc. Natl. Acad. Sci. U S A 2020, 117, 1009–1014. 10.1073/pnas.1919114117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.; Fan X.; Yan N. Cryo-EM Analysis of a Membrane Protein Embedded in the Liposome. Proc. Natl. Acad. Sci. U S A 2020, 117, 18497–18503. 10.1073/pnas.2009385117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Liu Y.; Yu Z.; Li S.; Feng S.; Cheng Y.; Agard D. A. General and Robust Covalently Linked Graphene Oxide Affinity Grids for High-Resolution Cryo-EM. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 24269–24273. 10.1073/pnas.2009707117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote L. J.; Kim F.; Huang J. Langmuir-Blodgett Assembly of Graphite Oxide Single Layers. J. Am. Chem. Soc. 2009, 131, 1043–1049. 10.1021/ja806262m. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Sengupta N.; Dutta S. Simplified Approach for Preparing Graphene Oxide TEM Grids for Stained and Vitrified Biomolecules. Nanomaterials 2021, 11, 643. 10.3390/nano11030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig A. C.; Frederick C. A.; Lippard S. J.; Nordlund P. Crystal Structure of a Bacterial Non-Haem Iron Hydroxylase That Catalyses the Biological Oxidation of Methane. Nature 1993, 366, 537–543. 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- Kopp D. A.; Gassner G. T.; Blazyk J. L.; Lippard S. J. Electron-Transfer Reactions of the Reductase Component of Soluble Methane Monooxygenase from Methylococcus Capsulatus (Bath). Biochemistry 2001, 40, 14932–14941. 10.1021/bi015556t. [DOI] [PubMed] [Google Scholar]
- Paulsen K. E.; Liu Y.; Fox B. G.; Lipscomb J. D.; Munck E.; Stankovich M. T. Oxidation-Reduction Potentials of the Methane Monooxygenase Hydroxylase Component from Methylosinus Trichosporium OB3b. Biochemistry 1994, 33, 713–722. 10.1021/bi00169a013. [DOI] [PubMed] [Google Scholar]
- Lee S. J.; McCormick M. S.; Lippard S. J.; Cho U. S. Control of Substrate Access to the Active Site in Methane Monooxygenase. Nature 2013, 494, 380–384. 10.1038/nature11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkx M.; Lippard S. J. Why OrfY? Characterization of MMOD, a Long Overlooked Component of the Soluble Methane Monooxygenase from Methylococcus Sapsulatus (Bath). J. Biol. Chem. 2002, 277, 5858–5865. 10.1074/jbc.M107712200. [DOI] [PubMed] [Google Scholar]
- Kim H.; An S.; Park Y. R.; Jang H.; Yoo H.; Park S. H.; Lee S. J.; Cho U. S. MMOD-Induced Structural Changes of Hydroxylase in Soluble Methane Monooxygenase. Sci. Adv. 2019, 5, eaax0059 10.1126/sciadv.aax0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazinsky M. H.; Lippard S. J. Correlating Structure with Function in Bacterial Multicomponent Monooxygenases and Related Diiron Proteins. Acc. Chem. Res. 2006, 39, 558–566. 10.1021/ar030204v. [DOI] [PubMed] [Google Scholar]
- Sirajuddin S.; Rosenzweig A. C. Enzymatic Oxidation of Methane. Biochemistry 2015, 54, 2283–2294. 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R.; Jones J. C.; Lipscomb J. D. Soluble Methane Monooxygenase. Annu. Rev. Biochem. 2019, 88, 409–431. 10.1146/annurev-biochem-013118-111529. [DOI] [PubMed] [Google Scholar]
- Suk J. W.; Kitt A.; Magnuson C. W.; Hao Y.; Ahmed S.; An J.; Swan A. K.; Goldberg B. B.; Ruoff R. S. Transfer of CVD-Grown Monolayer Graphene onto Arbitrary Substrates. ACS Nano 2011, 5, 6916–6924. 10.1021/nn201207c. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Lin L.; Sun L.; Huang Y.; Koh A. L.; Dang W.; Yin J.; Wang M.; Tan C.; Li T.; Tan Z.; Liu Z.; Peng H. Clean Transfer of Large Graphene Single Crystals for High-Intactness Suspended Membranes and Liquid Cells. Adv. Mater. 2017, 29, 1700639. 10.1002/adma.201700639. [DOI] [PubMed] [Google Scholar]
- Lin L.; Zhang J.; Su H.; Li J.; Sun L.; Wang Z.; Xu F.; Liu C.; Lopatin S.; Zhu Y.; Jia K.; Chen S.; Rui D.; Sun J.; Xue R.; Gao P.; Kang N.; Han Y.; Xu H. Q.; Cao Y.; Novoselov K. S.; Tian Z.; Ren B.; Peng H.; Liu Z. Towards Super-Clean Graphene. Nat. Commun. 2019, 10, 1912. 10.1038/s41467-019-09565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C.; Liu J.; Ma J.; Tan Z.; Zhang H. Stabilizing Effect of Oxygen on the Initial Stages of Poly(methyl methacrylate) Degradation. J. Therm. Anal. Calorim. 2016, 123, 1459–1467. 10.1007/s10973-015-5018-x. [DOI] [Google Scholar]
- Pathak M.; Kweon H.; Deo M.; Huang H. Kerogen Swelling and Confinement: Its implication on Fluid Thermodynamic Properties in Shales. Sci. Rep. 2017, 7, 12530. 10.1038/s41598-017-12982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Sun L.; Jia K.; Liu X.; Cheng T.; Peng H.; Lin L.; Liu Z. New Growth Frontier: Superclean Graphene. ACS Nano 2020, 14, 10796–10803. 10.1021/acsnano.0c06141. [DOI] [PubMed] [Google Scholar]
- Elango M.; Thamilselvan M.; Hemalatha V. In Sangameswari Structural and Optical Characterization on ZnS: Mn Filled PMMA Nanocomposites, International Conference on Nanoscience, Engineering and Technology (ICONSET 2011), 28–30 Nov, 2011; pp 11–13.
- Zhang Y. J.; Ren W.; Jiang Z. D.; Yang S. M.; Jing W. X.; Shi P.; Wu X. Q.; Ye Z. G. Low-Temperature Remote Plasma-Enhanced Atomic Layer Deposition of Graphene and Characterization of Its Atomic-Level Structure. J. of Mater. Chem. C 2014, 2, 7570–7574. 10.1039/C4TC00849A. [DOI] [Google Scholar]
- Psilodimitrakopoulos S.; Orekhov A.; Mouchliadis L.; Jannis D.; Maragkakis G. M.; Kourmoulakis G.; Gauquelin N.; Kioseoglou G.; Verbeeck J.; Stratakis E. Optical Versus Electron Diffraction Imaging of Twist-Angle in 2D Transition Metal Dichalcogenide Bilayers. Npj 2d Mater. Appl. 2021, 5, 77. 10.1038/s41699-021-00258-5. [DOI] [Google Scholar]
- Punjani A.; Rubinstein J. L.; Fleet D. J.; Brubaker M. A. cryoSPARC: Algorithms for Rapid Unsupervised Cryo-EM Structure Determination. Nat. Methods 2017, 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Tan P. H.; Han W. P.; Zhao W. J.; Wu Z. H.; Chang K.; Wang H.; Wang Y. F.; Bonini N.; Marzari N.; Pugno N.; Savini G.; Lombardo A.; Ferrari A. C. The Shear Mode of Multilayer Graphene. Nat. Mater. 2012, 11, 294–300. 10.1038/nmat3245. [DOI] [PubMed] [Google Scholar]
- Ferrari A. C.; Basko D. M. Raman Spectroscopy as a Versatile Tool for Studying the Properties of Graphene. Nat. Nanotechnol. 2013, 8, 235–246. 10.1038/nnano.2013.46. [DOI] [PubMed] [Google Scholar]
- Wu J. B.; Zhang X.; Ijas M.; Han W. P.; Qiao X. F.; Li X. L.; Jiang D. S.; Ferrari A. C.; Tan P. H. Resonant Raman Spectroscopy of Twisted Multilayer Graphene. Nat. Commun. 2014, 5, 5309. 10.1038/ncomms6309. [DOI] [PubMed] [Google Scholar]
- Lin Z.; Ye X.; Han J.; Chen Q.; Fan P.; Zhang H.; Xie D.; Zhu H.; Zhong M. Precise Control of the Number of Layers of Graphene by Picosecond Laser Thinning. Sci. Rep. 2015, 5, 11662. 10.1038/srep11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkx M.; Kopp D. A.; Sazinsky M. H.; Blazyk J. L.; Muller J.; Lippard S. J. Dioxygen Activation and Methane Hydroxylation by Soluble Methane Monooxygenase: A Tale of Two Irons and Three Proteins. Angew. Chem., Int. Ed. Engl. 2001, 40, 2782–2807. . [DOI] [PubMed] [Google Scholar]
- Whittington D. A.; Lippard S. J. Crystal Structures of the Soluble Methane Monooxygenase Hydroxylase from Methylococcus Capsulatus (Bath) Demonstrating Geometrical Variability at the Dinuclear Iron Active Site. J. Am. Chem. Soc. 2001, 123, 827–838. 10.1021/ja003240n. [DOI] [PubMed] [Google Scholar]
- Whittington D. A.; Rosenzweig A. C.; Frederick C. A.; Lippard S. J. Xenon and Halogenated Alkanes Track Putative Substrate Binding Cavities in the Soluble Methane Monooxygenase Hydroxylase. Biochemistry 2001, 40, 3476–3482. 10.1021/bi0022487. [DOI] [PubMed] [Google Scholar]
- Bepler T.; Kelley K.; Noble A. J.; Berger B. Topaz-Denoise: General Deep Denoising Models for CryoEM and CryoET. Nat. Commun. 2020, 11, 5208. 10.1038/s41467-020-18952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V.; Grosse-Kunstleve R. W.; Echols N.; Headd J. J.; Moriarty N. W.; Mustyakimov M.; Terwilliger T. C.; Urzhumtsev A.; Zwart P. H.; Adams P. D. Towards Automated Crystallographic Structure Refinement with Phenix.Refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Imprima E.; Floris D.; Joppe M.; Sanchez R.; Grininger M.; Kuhlbrandt W. Protein Denaturation at the Air-Water Interface and How to Prevent it. Elife 2019, 8, e42747 10.7554/eLife.42747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-Time CTF Determination and Correction. J. Struct. Biol. 2016, 193, 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig A. C.; Brandstetter H.; Whittington D. A.; Nordlund P.; Lippard S. J.; Frederick C. A. Crystal Structures of the Methane Monooxygenase Hydroxylase from Methylococcus Capsulatus (Bath): Implications for Substrate Gating and Component Interactions. Proteins 1997, 29, 141–152. . [DOI] [PubMed] [Google Scholar]
- Sazinsky M. H.; Lippard S. J. Product Bound Structures of the Soluble Methane Monooxygenase Hydroxylase from Methylococcus Capsulatus (Bath): Protein Motion in the Alpha-subunit. J. Am. Chem. Soc. 2005, 127, 5814–5825. 10.1021/ja044099b. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera-a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N. Automated Electron Microscope Tomography Using Robust Prediction of Specimen Movements. J. Struct. Biol. 2005, 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Hagen W. J. H.; Wan W.; Briggs J. A. G. Implementation of a Cryo-Electron Tomography Tilt-Scheme Optimized for high Resolution Subtomogram Averaging. J. Struct. Biol. 2017, 197, 191–198. 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. R.; Mastronarde D. N.; McIntosh J. R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Chen M.; Dai W.; Sun S. Y.; Jonasch D.; He C. Y.; Schmid M. F.; Chiu W.; Ludtke S. J. Convolutional Neural Networks for Automated Annotation of Cellular Cryo-Electron Tomograms. Nat. Methods. 2017, 14, 983–985. 10.1038/nmeth.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Meng E. C.; Couch G. S.; Croll T. I.; Morris J. H.; Ferrin T. E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Eevelopers. Protein Sci. 2021, 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A.; Eisenbraun B.; Key J.; Sanschagrin P. C.; Timony M. A.; Ottaviano M.; Sliz P. Collaboration Gets the Most Out of Software. Elife 2013, 2, e01456 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.