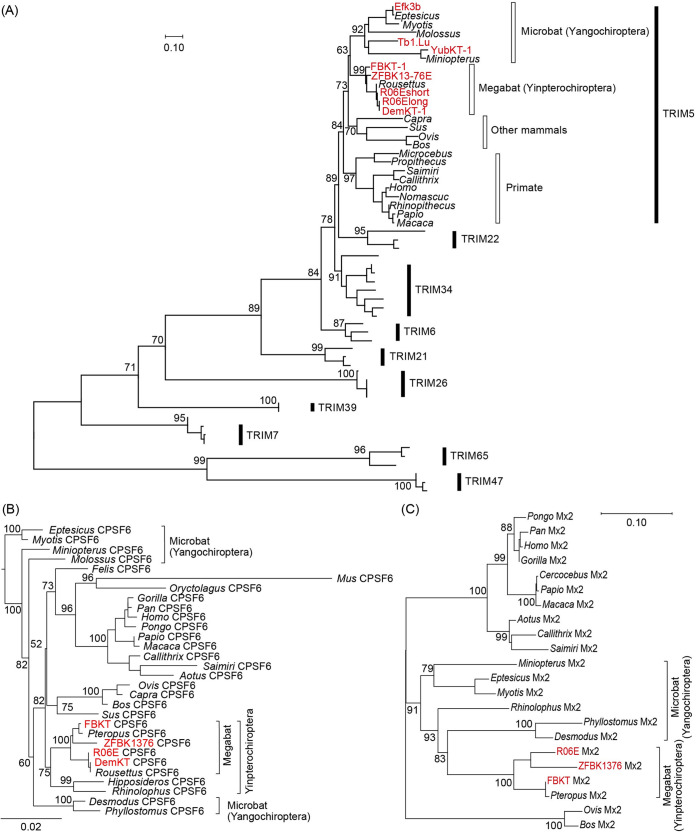
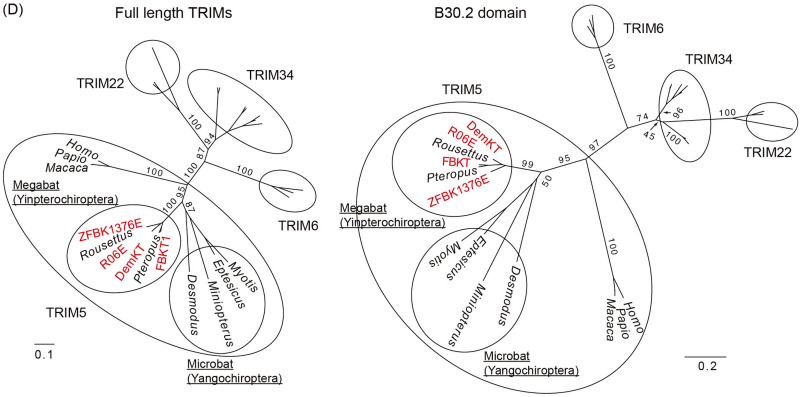
FIG 5.
Phylogeny of newly isolated megabat CPSF6, Mx2/MxB, and TRIM5α. (A) An ML phylogenetic tree of the newly identified TRIM5α (red) and those of other mammals based on the RBCC domain of C-IV type TRIMs. Numbers at the nodes of the main clusters indicate the bootstrap percentage (%) generated from 1,000 repeats. The new bat TRIMs formed a clade with TRIM5 of various mammalian species (bootstrapping value = 84%). (B) An ML phylogenetic tree of CPSF6 genes of 30 mammalian species. The newly cloned pteropid bat CPSF6 (highlighted in red) formed a clade with the Pteropus and Rousettus CPSF6 obtained from NCBI GenBank (bootstrapping value = 100%). We estimated the dN/dS ratio at each branch using codeml (see Materials and Methods), and the dN/dS ranged from 0.0001 to 0.0911, suggesting a negative selection of the CPSF6 gene. (C) A phylogenetic tree of pteropid megabat and other mammalian Mx2. The newly identified R06E Mx2 is highlighted in red. Due to the loss of almost the entire 3′ half (Fig. S1D), DemKT-1 Mx2/MxB was not included in the phylogenetic analysis. (D) Unrooted phylogenetic ML trees of TRIM5α and its close relatives (TRIM34, TRIM6, and TRIM22) based on the full-length (left) or the B30.2 domain (right) sequences. Newly identified megabat TRIM5α are shown in red, and the Yinpterochiroptera and Yangochiroptera clades are highlighted in ovals in the TRIM5α clade. For both phylogenetic trees, the numbers at the nodes of each main cluster indicate the bootstrap values generated from 1,000 repeats. GenBank accession numbers of the sequences used in each tree are provided in Table S1.