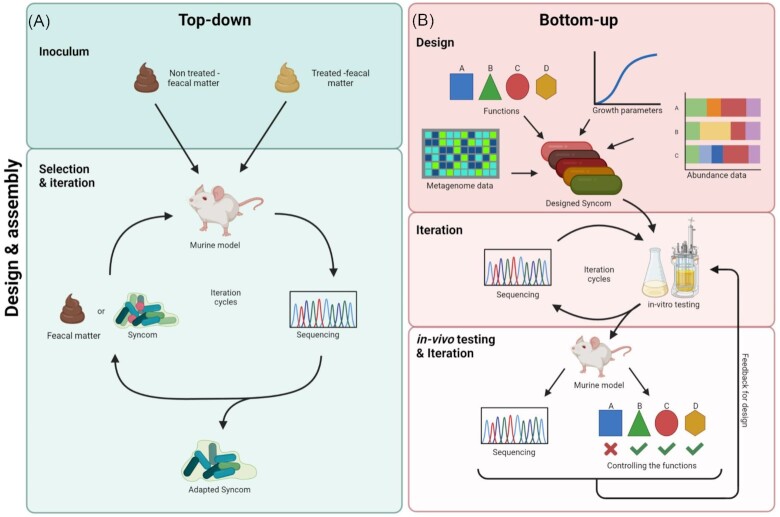
Figure 2.
Overview on the strategies for designing, assembling, and testing SynComs. (A) The top-down approach starts at the inoculum and microbial composition profiling, followed by the selection and iteration cycle in which bacteria are added to an animal model, isolated, and subsequently sequenced for a further iteration cycle or ultimately the description of an adapted SynCom (Atarashi et al. 2013). (B) The bottom-up approach uses existing knowledge on the metagenome, abundances, and growth parameters of candidate microorganisms. Furthermore, when the SynCom is required to have a specific function, the candidate microorganisms can be adjusted accordingly (van der Lelie et al. 2021). The designed SynCom then enters iteration cycles in the in-vitro setting, being cultured in batches and sequenced to understand the composition. Lastly, the SynCom is tested in an in-vivo setting, being introduced to a murine model. The SynCom is then extracted from the model and sequenced after an incubation or experimental period. The functional profile is also checked to ensure the SynCom performed as expected. From there, feedback from the results can be used to start the iteration cycles again.