Fig. 1.
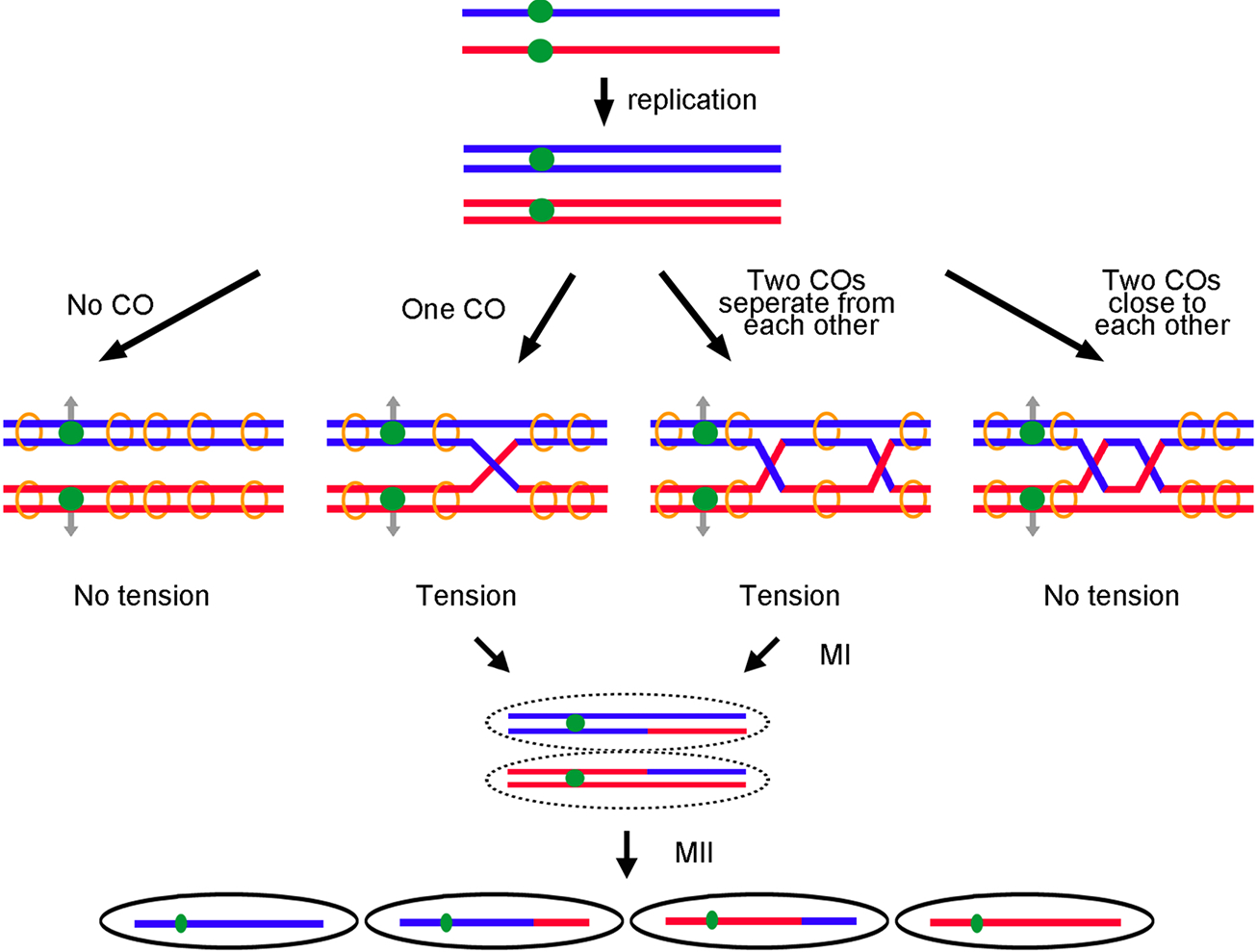
Crossovers are needed for proper chromosome segregation in meiosis. Red and blue lines indicate replicated homologs from each parent. Each line indicates a double-stranded DNA of one chromatid. Green dots represent centromeres. After replication, sister chromatids are held together by cohesins (orange circles). Gray arrows indicate the direction of homologous centromere separation at the first meiotic division (MI). (Left panel) No tension is generated between homologs when no CO is formed. (Middle two panels) Tension is formed between homologs when one CO or two well-separated COs arise. (Right panel) Two COs too close to each other may have too little sister chromatid cohesion between the crossovers to produce tension between homologs to aid segregation. The tension ensures homologous centromeres segregate properly in MI. Crossovers also generate recombinant haploid gametes after the second meiotic division (MII).
