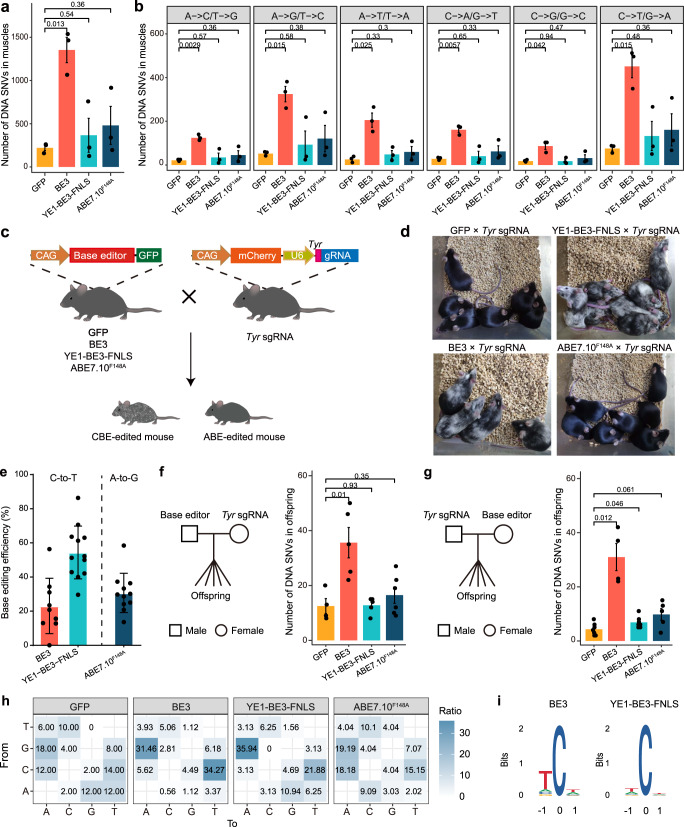
Fig. 2. Off-target DNA mutations in the transgenic mice expressing base editors.
a Comparison of the total number of DNA SNVs in muscles of GFP, BE3 YE1-BE3-FNLS and ABE7.10F148A transgenic mice. n = 3 for each group. b Comparison of the number of DNA SNVs with the indicated mutation types in muscles of the GFP, BE3, YE1-BE3-FNLS, and ABE7.10F148A transgenic mice. n = 3 for each group. c Schematic diagram for in vivo base editing verification by crossing Tyr sgRNA transgenic mice with GFP, BE3, YE1-BE3-FNLS, and ABE7.10F148A mice. d Representative photos for the hair colors of offspring from GFP × Tyr sgRNA, BE3 × Tyr sgRNA, YE1-BE3-FNLS × Tyr sgRNA and ABE7.10F148A × Tyr sgRNA mice. e The C-to-T or A-to-G base editing efficiency at Tyr target site in offspring of BE3 × Tyr sgRNA (n = 9), YE1-BE3-FNLS × Tyr sgRNA (n = 12) or ABE7.10F148A × Tyr sgRNA (n = 11) breeding pairs. f Comparison of the number of de novo SNVs in offspring expressing GFP (n = 4), BE3 (n = 5), YE1-BE3-FNLS (n = 5) or ABE7.10F148A (n = 6) with the Tyr sgRNA transgenic mice as mother. g Comparison of the number of de novo SNVs in offspring expressing GFP (n = 7), BE3 (n = 4), YE1-BE3-FNLS (n = 7) or ABE7.10F148A (n = 5) with the Tyr sgRNA transgenic mice as father. n = biologically independent samples. Data are presented as mean ± SEM. h Distribution of mutation types of the de novo SNVs in offspring of crossing GFP/BE3/YE1-BE3-FNLS/ABE7.10F148A father and Tyr sgRNA mother. The number indicates the percentage of a certain type of mutation among all mutations. i Sequence logos derived from the de novo SNVs in offspring of crossing BE3/YE1-BE3-FNLS father and Tyr sgRNA mother. Source data are provided as a Source Data file.