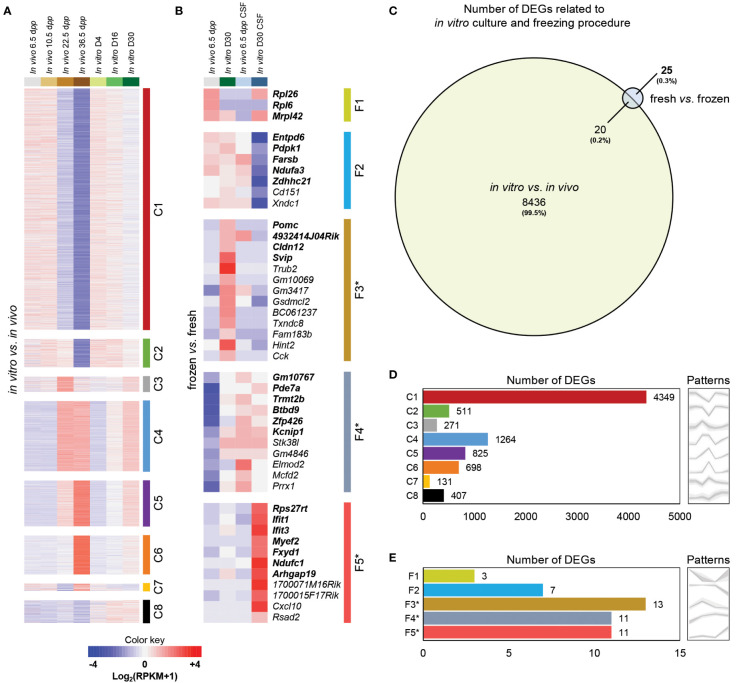
Figure 2.
Global gene expression profiling of testicular tissue samples in response to in vitro culture or freezing procedures. False-color heatmap summarizing the patterns defining the global concentrations for transcripts across the entire sample set focused on the impact of in vitro culture (A) and freezing procedure (B). Each row corresponds to the mean of biological replicate samples: in vivo 6.5 (fresh: n=3; CSF: n=2), 10.5 (n=3), 22.5 (n=3), 36.5 (n=2) dpp; in vitro D4 (n=3), D16 (n=2), and D30 (fresh tissue: n=3; CSF: n=1). C1 to C8 and F1 to F5* refer to the individual patterns for the impact of in vitro culture and freezing procedure, respectively. DEGs only present for fresh vs. frozen are in bold. Fold change values are shown with a red or blue background indicating up-regulation or down-regulation, respectively. A color key-scale is shown for standardized values in Log2(RPKM+1). (C) Area-proportional Venn Diagram showing the overlap of significantly DEGs in in vitro vs. in vivo (with 8436 uniquely mapped transcripts from a total of 8456 DEGs) compared to fresh vs. frozen (with only 25 uniquely mapped transcripts from a total of 45 DEGs). Numbers labeled in each part show the amount of the genes with the corresponding expression patterns. The number of significantly DEGs included in each pattern are shown for the impact of in vitro culture (D) and freezing procedure (E). Grey patterns are the graphical visualization of the average expression levels of the several patterns (with the variations between repetitions). F3* (23.1%), F4* (18.2%), and F5* (18.2%) were patterns with a large proportion of DEGs starting with ‘Gm-’ or ending with ‘Rik’ who are predicted genes or annotated genes that do not have a canonical name (yet). Based on Ensembl biotype classifications in mouse (https://www.ensembl.org/Mus_musculus/Info/Index), genes starting with ‘Gm-’ or ending with ‘Rik’ include protein-coding RNAs, long non-coding RNAs, and antisense transcripts. Interestingly, genes with the ‘Gm’ prefix are enriched for pseudogenes. CSF, controlled slow freezing; DEGs, differentially expressed genes; dpp, days postpartum; RPKM, reads per kilobase million.