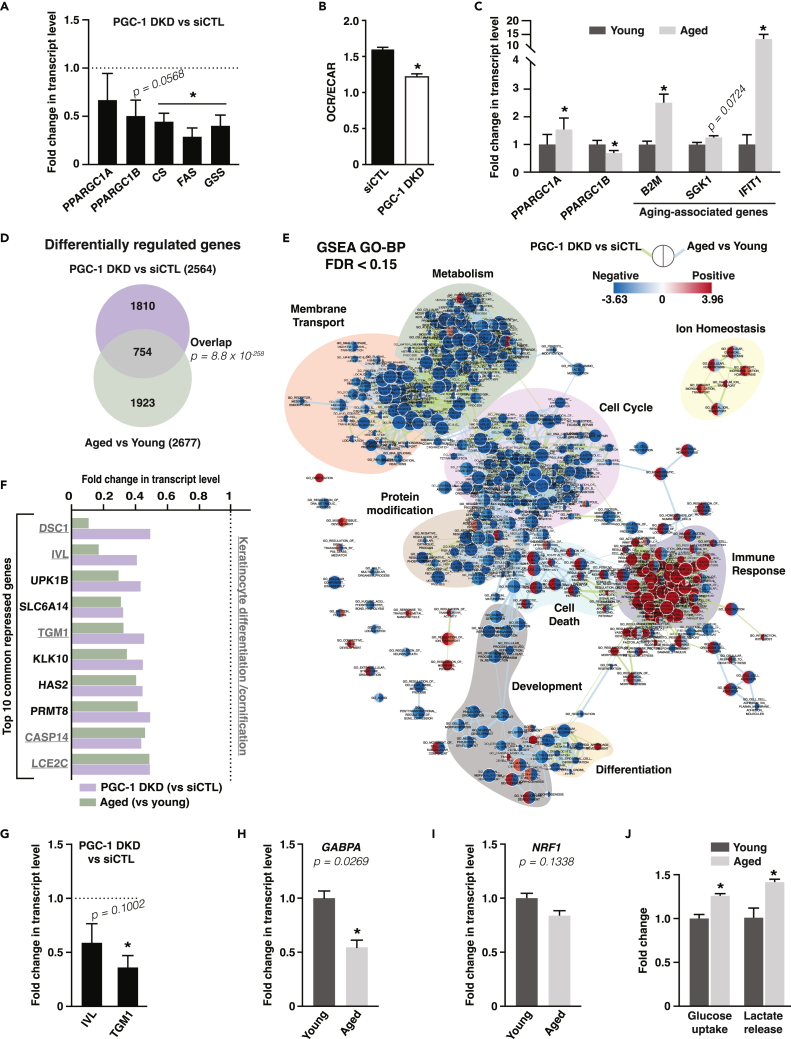
Figure 2.
PGC-1α and PGC-1β double silencing in human primary keratinocytes mimics aspects of keratinocyte aging and represses cornification and terminal differentiation gene programs
(A) Expression of PGC-1s and targets in normal human epidermal keratinocytes (NHEK) 3 days after PGC-1α/β double knockdown (PGC-1 DKD). Data are shown as mean +SEM from 4 independent experiments. ∗p < 0.05, paired Student’s t test.
(B) OCR/ECAR ratio of keratinocyte HaCaT cells transfected with control siRNA or PGC-1α/β double knockdown (PGC-1 DKD). Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(C) Gene expression analysis in NHEK from young (28 y.o.) and aged (83 y.o.) donors. Data are shown as mean + SEM from 6 independent experiments. ∗p < 0.05, paired Student’s t test.
(D) Venn diagram from microarray data analyses grouping differentially regulated genes upon PGC-1 DKD versus siCTL in NHEK and upon aging in NHEK from aged (83 y.o.) versus young (28 y.o.) donors. 754 genes are common to PGC-1s DKD and keratinocyte aging and are analyzed in panels E-F.
(E) Network analysis of enriched and depleted pathways upon PGC-1 DKD versus siCTL in NHEK and upon aging in NHEK from aged (83 y.o.) versus young (28 y.o.) donors. Enrichment (red) or depletion (blue) of each pathway is shown in nodes, where the left side represents PGC-1 DKD versus siCTL and the right represents aged versus young. Edges link pathways (nodes) with common genes, and edge length is inversely proportional to the similarity between nodes. Color-shaded pathway groups were outlined manually.
(F) Top 10 common genes repressed upon PGC-1 DKD and keratinocyte aging. Underlined transcripts are known to be involved in the differentiation or cornification processes. Fold change is from transcriptome array analysis. Dotted line corresponds to control keratinocytes from which fold changes are calculated.
(G) Validation of the repression of terminal differentiation markers genes (IVL: involucrin; TGM1: transglutaminase 1) upon PGC-1 DKD in NHEK by real-time PCR. Data are shown as mean + SEM from 4 independent experiments. ∗p < 0.05, paired Student’s t test.
(H) Expression of GABPA in NHEK from aged (83 y.o.) versus young (28 y.o.) donors, normalized to the expression of NHEK cells from young donor. Data are shown as mean + SEM from 5 independent experiments. ∗p < 0.05, paired Student’s t test.
(I) Expression of NRF1 in NHEK from aged (83 y.o.) versus young (28 y.o.) donors, normalized to the expression of NHEK cells from young donor. Data are shown as mean + SEM from 6 independent experiments. Paired Student’s t test.
(J) Glucose uptake and lactate release in NHEK from aged (83 y.o.) versus young (28 y.o.) donors, normalized to the expression of NHEK cells from young donor. Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.