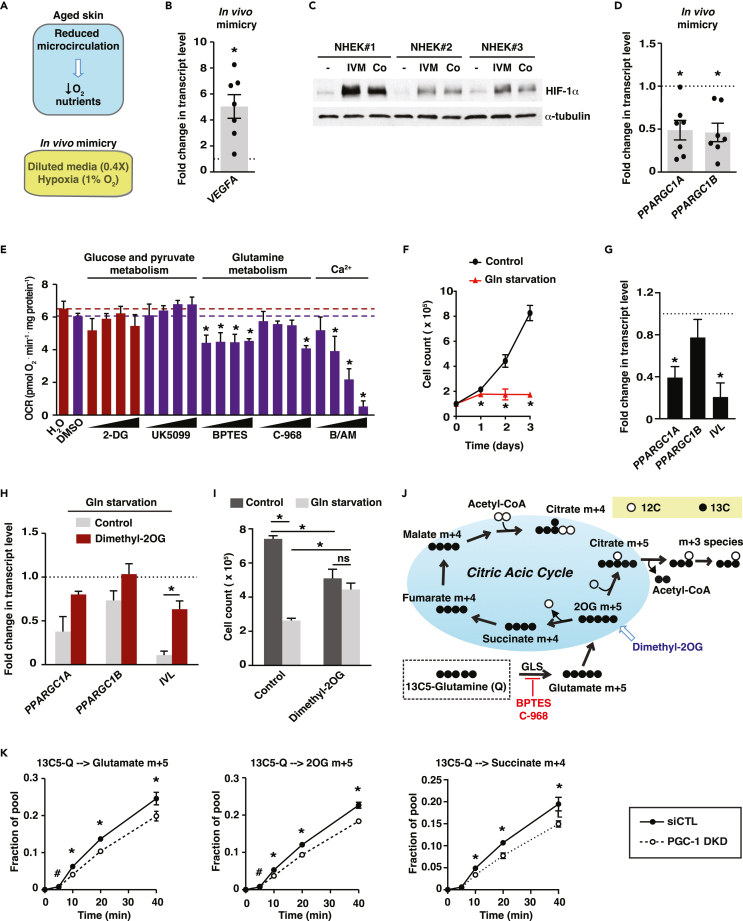
Figure 4.
Crosstalk between glutamine availability and PGC-1s in human keratinocytes
(A) Schematic depicting reduced microcirculation accompanied by lowered levels of oxygen and nutrients in aged skin. This was mimicked experimentally by growing cells in 1% oxygen and diluted isotonic media (0.4X).
(B) VEGFA gene expression in normal human epidermal keratinocytes (NHEK) subjected to microenvironment mimicry (1% hypoxia and diluted nutrients) for 24 h. Data are shown as fold change compared with normoxic and full nutrient conditions (dotted line) and as mean ± SEM. NHEK were from 7 different donors (each dot), each analyzed one or two times independently. ∗p < 0.05, paired Student’s t test.
(C) Immunoblot analyses for HIF-1α in protein extracts from NHEK exposed to low oxygen and nutrient conditions (IVM: in vivo mimicry, as described in B). NHEK were from 3 different donors. Cobalt chloride (Co, 200 μM) was used as positive control for HIF-1α induction. α-tubulin was used as loading control.
(D) PPARGC1A and PPARGC1B gene expression in NHEK cells subjected to microenvironment mimicry as in B for 24 h. Data are shown as fold change compared with normoxic and full-nutrient conditions (dotted line) and as mean ± SEM. NHEK were from 7 different donors (each dot), each analyzed one or two times independently. ∗p < 0.05, paired Student’s t test.
(E) Oxygen consumption rate (OCR) of keratinocyte HaCaT cells treated for 2 days with 4 doses of various compounds: 2-DG: 2-deoxyglucose (0.5, 1, 2, 4 mM), UK5099 (1, 3, 10, 30 μM), BPTES (1, 3, 10, 30 μM), C-968: compound 968 (0.3, 1, 3 10 μM) and B/AM: BAPTA/AM (3, 10, 20, 30 μM). Treatments were compared to specific control treatment (dotted line) to which they are associated by a color code (H2O: red, DMSO: purple). Data are expressed as mean + SEM from 3 independent experiments. ∗p < 0.05, one-way ANOVA, Dunnett’s post-test.
(F) Proliferation curves of keratinocyte HaCaT cells under 4 mM glutamine or glutamine starvation. Data are shown as mean ± SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(G) Gene expression analyses of glutamine-starved (3 days) keratinocyte HaCaT cells. Data are shown as fold change versus control conditions (4 mM glutamine) and mean + SEM from 7 independent experiments. ∗p < 0.05, paired Student’s t test.
(H) Dimethyl-2OG (4 mM) rescues gene expression of glutamine-starved keratinocyte HaCaT cells. Data are expressed as fold change versus control conditions (4 mM glutamine) and as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(I) Dimethyl-2OG (4 mM) rescues the proliferation inhibition of glutamine-starved keratinocyte HaCaT cells. Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, two-way ANOVA, Tukey post-test.
(J) Schematic showing the metabolism of 13C5-L-glutamine in the citric acid cycle.
(K) Stable isotope tracer analysis using 13C5-L-glutamine in HaCaT cells treated with siCTL or PGC-1 DKD. Dynamic labeling of glutamate M+5, alpha-ketoglutarate (2OG) M+5, and succinate M+4 are shown. Data are shown as mean ± SEM from 3 independent experiments. ∗p < 0.05, #p < 0.10, paired Student’s t test.