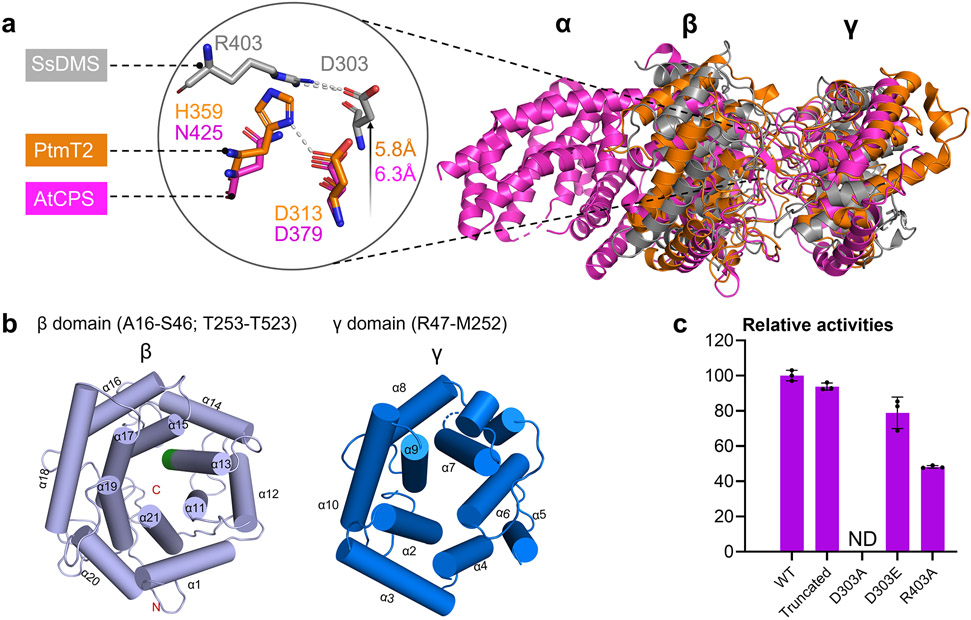
Figure 3. The apo-structure of SsDMS.
(a) Overall structural comparison of apo-SsDMS (PDB ID: 7XQ4, gray), AtCPS (PDB ID: 4LIX, magenta) and PtmT2 (PDB ID: 5BP8, orange). The location of catalytic acid group of D303 and R403 in SsDMS is relatively higher than that in PtmT2 and AtCPS. (b) β- (light blue helices) and γ-domain (marine blue helices) in SsDMS. The conserved catalytic DxDD motif in β-domain is located in α13 helice and shown in green; the dashed loop in γ-domain represents the disordered residues R160–Q163. (c) Relative enzyme activities of D303A, D303E, and R403A mutants, along with the truncated SsDMS used in crystallization. ND, not detected. Error bars indicate the standard deviation of three independent replicates.