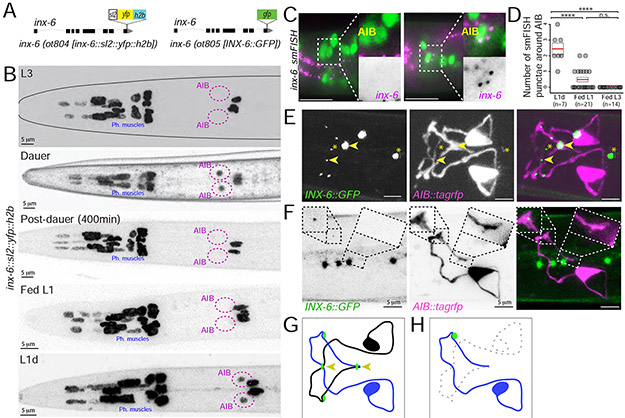
Fig. 3: Dauer-induced expression of inx-6 in AIB generates new gap junctions with che-7.
(A) Schematics of inx-6 transcriptional reporter allele, inx-6(ot804) and GFP-tagged inx-6 translational reporter allele, inx-6(ot805).
(B) inx-6 reporter allele, inx-6(ot804) is additionally turned on in AIB interneurons in dauer that disappears in post-dauer stage. inx-6 allele is also expressed in AIB in L1d.
(C) smFISH against endogenous inx-6 mRNA (magenta) in Fed-L1 and L1d. AIB was marked by eat-4::yfp (otIs388) expression (green). Inset shows enlargement of AIB.
(D) Quantification of inx-6 smFISH data. Each circle represents the number of AIB associated smFISH puncta in a single animal, red lines indicate the mean and rectangles indicate S.E.M. Wilcoxon rank-sum tests p-values (n.s. = non significant and ****p<0.0001).
(E) Expression of INX-6 puncta (green), in inx-6(ot805) dauer. npr-9p::tagrfp (otIs643) expression marks AIB processes (magenta). Arrowheads mark INX-6::GFP at the crossover points of AIBL and AIBR. Asterisks mark INX-6 puncta in pharyngeal muscles.
(F) Expression of INX-6 puncta (green) on AIBL (magenta) in daf-7(e1372) dauers (inx-6(ot805); daf-7(e1372); otIs643), where AIBR was ablated. In absence of AIBR, INX-6 punctum at the AIBL-AIBR crossover point (right box) disappear, while has no effect on INX-6 puncta in other region (left box). See Fig. 3E for control (pre-ablation) image. Four INX-6 puncta that do not overlap with AIB-processes are among pharyngeal muscles.
(G) Schematic of major INX-6 puncta (green circles) on AIB. Arrowheads mark INX-6 puncta at the AIBL-AIBR crossover points.
(H) Schematic of the effect of AIBR-ablation on INX-6 puncta on the remaining AIBL neuron. See Fig. 3G for control (pre-ablation) schematic.