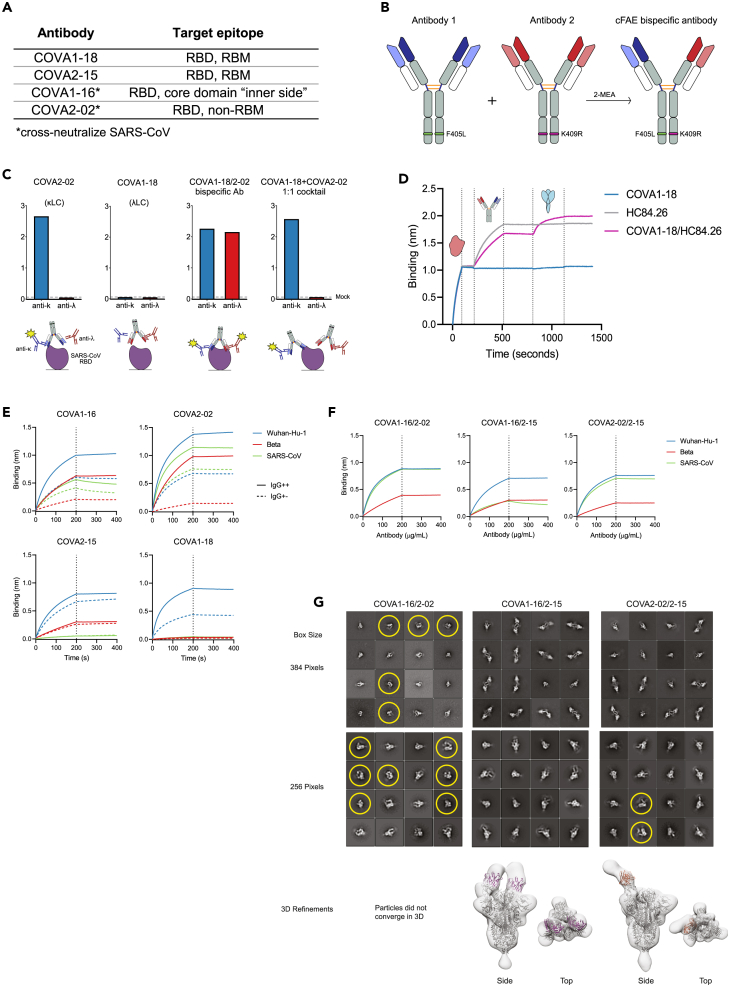
Figure 1.
Characterization of IgG1-based bsAbs that bind SARS-COV-2 and SARS-COV S proteins
(A) Selected COVA NAbs and their target epitopes.
(B) Schematic depiction of the method used for bsAb production (cFAE). Matching point mutations (F405L and K409R; EU numbering) are introduced in the parental antibodies, which then undergo Fab arm exchange in the presence of reducing agent 2-MEA, forming IgG-like bsAbs.
(C) Confirmation of bispecificity by ELISA. His-tagged SARS-CoV RBD was bound to an nickel–nitrilotriacetic acid (NiNTA) ELISA plate, followed by the bsAbs or mAb controls and secondary Abs specific for either kappa (i.e. COVA-2-02) or lambda LC (i.e. COVA1-18). Detected signals are depicted in the schematic with yellow stars.
(D) “Sandwich” biolayer interferometry (BLI) traces to confirm concurrent bsAb binding. BsAb or mAb controls were added after his-tagged hepatitis C virus (HCV) E2 was loaded onto NiNTA biosensors. Subsequently, SARS-CoV-2 S was added to measure a second association.
(E) BLI sensorgrams of IgG++ and IgG+- formats of selected COVA NAbs binding to Wuhan-Hu-1, Beta, and SARS-CoV S proteins. The dotted lines represent the end of NAb association and the start of dissociation.
(F) BLI sensorgrams of bsAbs binding to Wuhan-Hu-1, Beta, and SARS-CoV S proteins. The curves are representative of two independent experiments.
(G) Representative 2D class averages from NS-EM analysis and corresponding 3D reconstructions, COVA1-16/2–15 and COVA2-02/2–15 bsAbs bound to SARS-CoV-2-6P-Mut7 spike protein. Due to heterogeneity, particles did not converge to a stable 3D class for COVA1-16/2-02. For the 2D classes, datasets were processed with a box size of 384 pixels to show inter-spike avidity and a box size of 256 pixels to show single spike proteins. Trimer degradation in the COVA1-16/2-02 and COVA2-02/2–15 complexes is highlighted with yellow circles. Fc portions of the bsAbs in the 2D classes are seen as faint ghost densities near the Fabs. For the 3D refinements, a box size of 256 pixels was used to focus on a single spike complex. PDB: 6VYB (one RBD-up) and a “dummy” (poly alanine) Fv model were fit into the maps.
See also Figure S1.