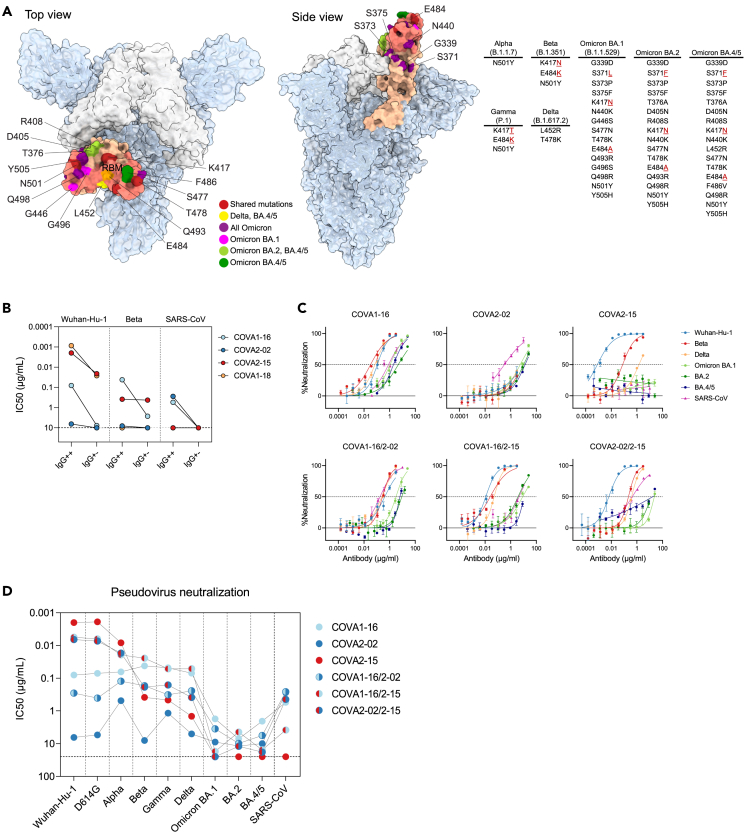
Figure 3.
BsAbs potently neutralize SARS-CoV-2 variants and SARS-CoV in pseudovirus neutralization assays
(A) Structure of SARS-CoV-2 S trimer with 2 “down” RBD’s (white) and one “up” (salmon) (PDB: 6ZGG). The RBM of the RBD is highlighted in darker salmon. RBD mutations of SARS-CoV-2 variants are listed in a table and pointed out on the RBD structure in red (positions shared between several variants), yellow (mutation shared between Delta and Omicron BA.4/5), purple (mutations shared between all Omicron subvariants), magenta (mutations unique to Omicron BA.1), light green (mutations shared between Omicron BA.2 and BA.4/5), and dark green (mutations unique to Omicron BA.4/5).
(B) Half maximal inhibitory concentration (IC50) values of COVA NAbs (IgG++) or “dead arm” bsAbs (IgG+-) against Wuhan-Hu-1, Beta and SARS-CoV pseudoviruses. In case of IC50 > 10 μg/mL (indicated by a dotted line) the antibody was considered non-neutralizing.
(C) Representative neutralization curves of monospecific and bispecific COVA NAbs against SARS-CoV-2 (Wuhan-Hu-1), SARS-CoV-2 variants and SARS-CoV pseudovirus neutralization. The dotted lines indicate 0% and 50% neutralization. Shown is the mean ± SEM of technical triplicates.
(D) IC50 titers of sarbecovirus neutralization by COVA monospecific and bispecific NAbs. In case of IC50 > 25 μg/mL (indicated by a dotted line) the antibody was considered non-neutralizing. Each dot represents the mean IC50 value of at least two independent experiments performed in triplicate. Connected dots represent data from the same NAb.
See also Figures S4 and S5.