Extended Data Fig. 8 |. Analysis of genome editing outcomes with CTS templates and small molecule inhibitors.
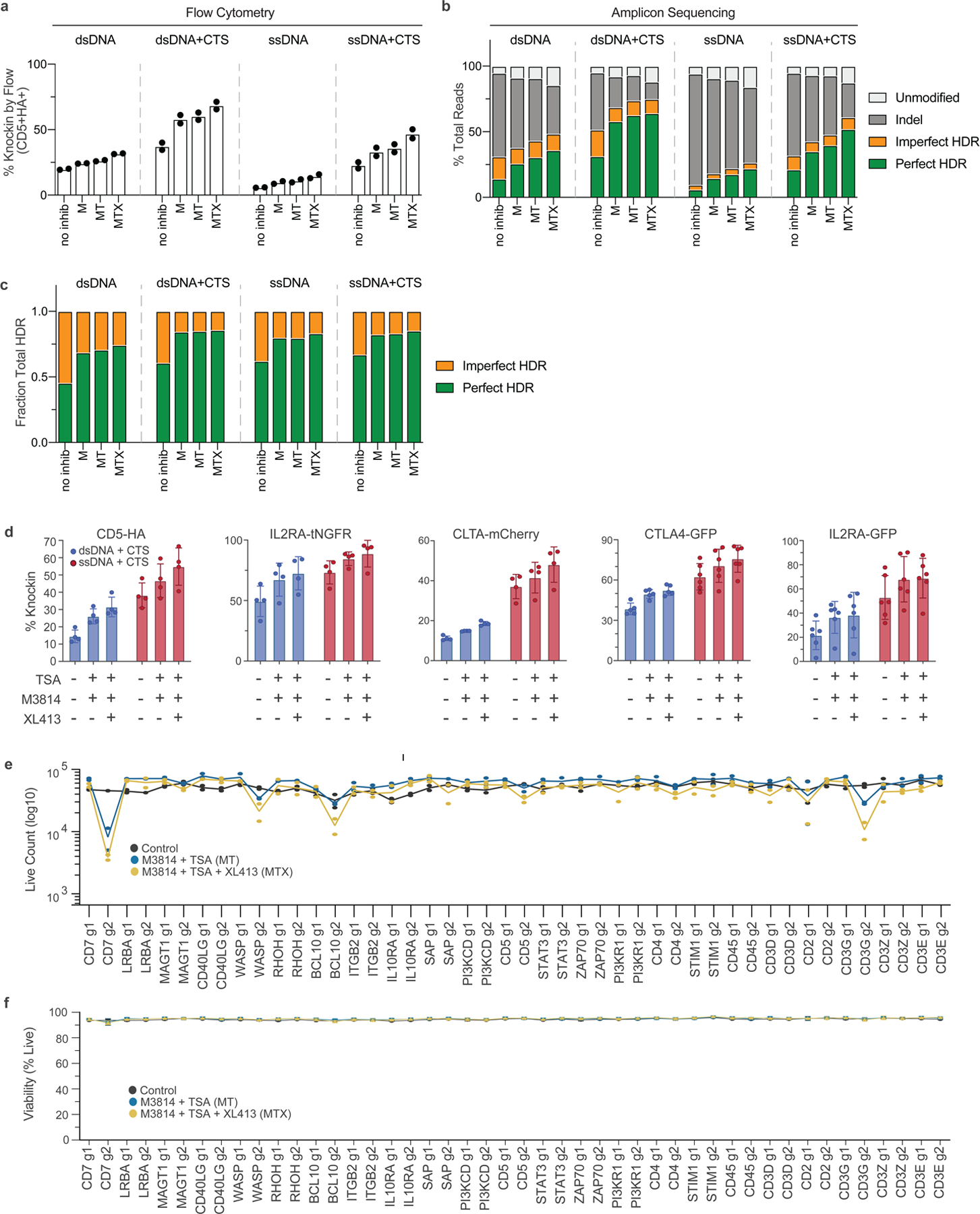
(a-c) Evaluation of genome editing outcomes by either (a) flow cytometry or (b) amplicon sequencing using small dsDNA, dsCTS, ssDNA, or ssCTS CD5-HA HDRTs at non-toxic concentrations (800 nM) with and without M, MT, and MTX inhibitor combinations. (c) Ratio of perfect:imperfect HDR events with each combination. (d) Comparison of dsCTS and ssCTS templates in combination with small molecular inhibitors in 5 different knock-in constructs using a large CD5-HA HDRT (−2.7 kb, n = 4 donors), a tNGFR knock-in to the IL2RA gene (~1.5 kb, n = 4 donors), an mCherry fusion in the clathrin gene (~1.5 kb, n = 4 donors), a near full length CTLA-4-GFP fusion to the CTLA4 gene (~2.1 kb, n = 6 donors), and a full length IL2RA-GFP fusion to the IL2RA gene (~2.3 kb, n = 6 donors). (e-f) Evaluation of (e) live cell counts and (f) viability +/− MT and MTX inhibitor combinations using 44 different knock-in constructs targeting a tNGFR marker across 22 different target loci with 2 gRNA per gene (g1 and g2). Panel a shows mean and individual values from two healthy blood donors. Panels b, c, e, and f show mean values from two healthy blood donors. Panel d shows mean +/− SD. CTS = Cas9 target site, HDRT = homology-directed repair template, dsCTS = dsDNA + CTS HDRT, ssCTS = ssDNA + CTS HDRT, M = M3814, MT = M3814 + TSA, MTX = M3814 + TSA + XL413.
