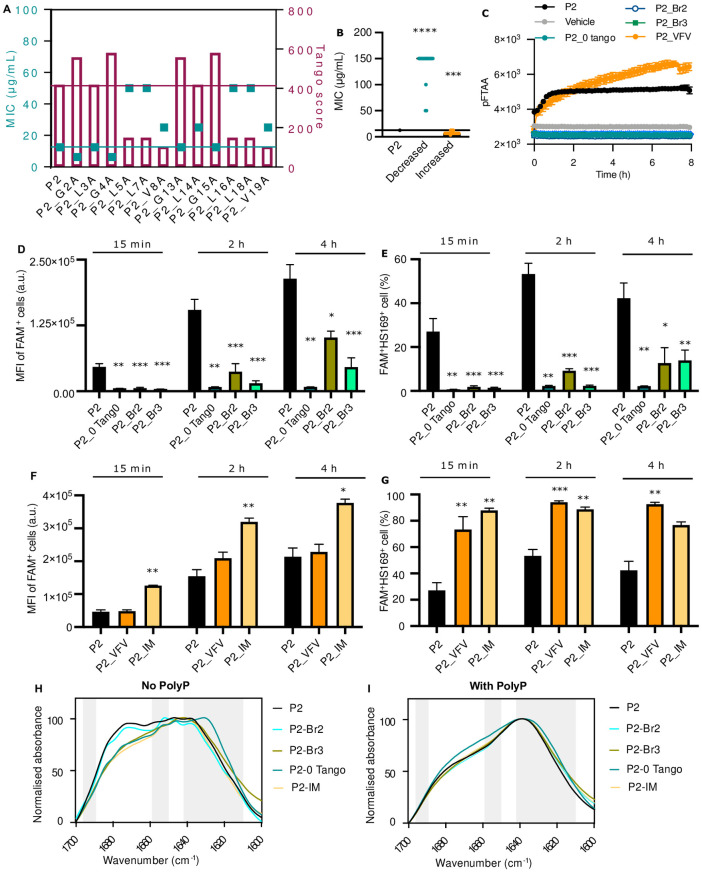
Fig 2. Increased aggregation propensity enhances Pept-in antimicrobial potency by promoting Pept-in accumulation and IB formation.
A: MIC (left Y-axis) against E. coli BL21 and Tango score (right Y-axis) of P2 derivatives generated by alanine scanning of the APR region. This figure is associated with data from S3 Table. The red bars represent the APR TANGO score, and the red line is the baseline of the P2 APR Tango score. The blue squares represent MIC values, and the blue line is the baseline of P2 MIC. B: MIC of P2 variants generated by aggregation propensity modification against E. coli BL21. This figure is associated with data from S5 Table. Each dot represents the MIC of one Pept-in design. C-G: P2 variants with decreased aggregation propensity: P2_0 Tango, P2_Br2, P2_Br_3; with increased aggregation propensity: P2_VFV, P2_IM. C: Time dependence of pFTAA (0.5 μM) fluorescence intensity of P2 derivatives (50 μM) in the presence of polyP (0.5 mM) (n = at least 6). D-G: Flow cytometry analysis of E. coli BL21 treated with FAM-labelled Pept-ins for different time points at FAM-P2-MIC from three independent experiments. Samples were acquired using Gallios Flow Cytometry. MFI of FAM+ cells (D), and the percentage+ of FAM and HS169+ cells (E) when treated with P2 and its derivatives with decreased antimicrobial potency. MFI of FAM+ cells (F), and the percentage+ of FAM and HS169+ cells (G) when treated with P2 and its derivatives with increased antimicrobial potency. Error bars represent SEM. H-I: FTIR spectrum of P2 variants with altered aggregation propensity in PBS (6% DMSO), without (H) or with (I) the presence of polyP. The absorbance is normalised between all samples and the spectrum is scaled to the amide I region between 1600–1700 cm−1. Peaks within the left (1689–1696 cm−1) and right (1610–1642 cm−1) grey bar are assigned to β-sheet, while peaks within the grey bar in the middle (1651–1659 cm−1) is assigned to α-helix. The FTIR spectrums are representative of three independent experiments. For B, one-sample Wilcoxon signed-rank test was used to compare the MIC median of each Pept-in group to P2 MIC (12.5 μg/mL). For D-G, a two-tailed Student t-test was performed for calculating statistical significance between the mean of P2 variant and P2 (n = at least 6). Asterisks indicating the level of the p value centred over the error bar mean: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.