Figure 4. Specificity profiling of tyrosine kinases using the pTyr-Var library.
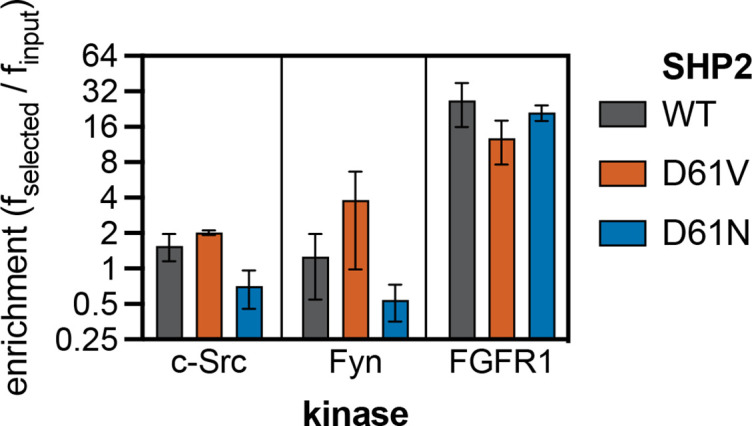
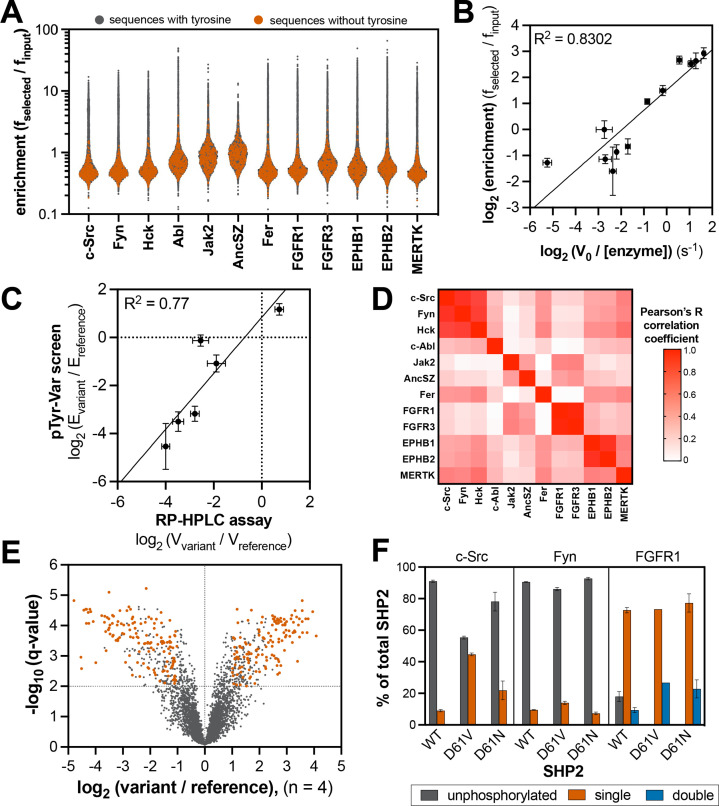
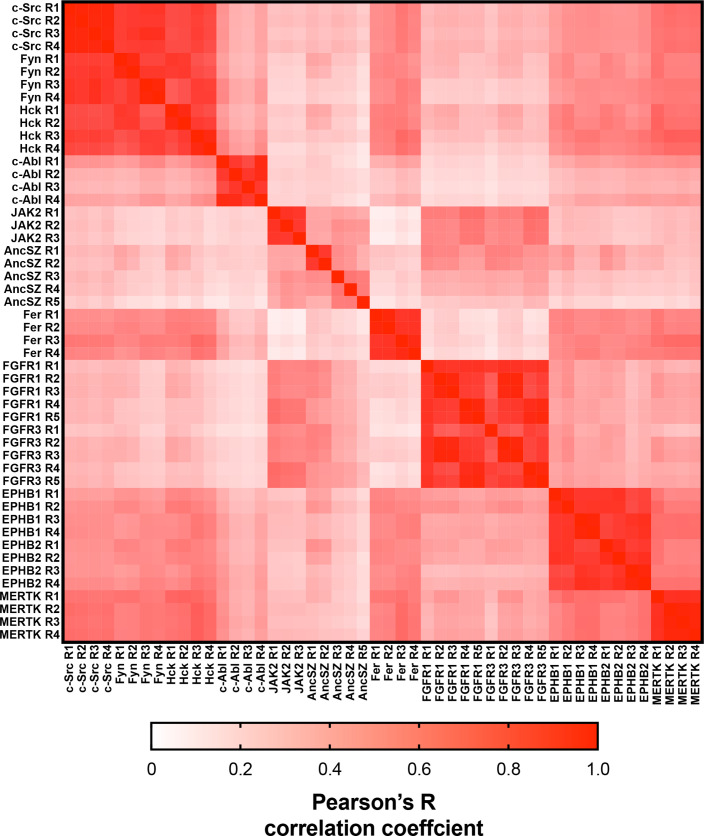
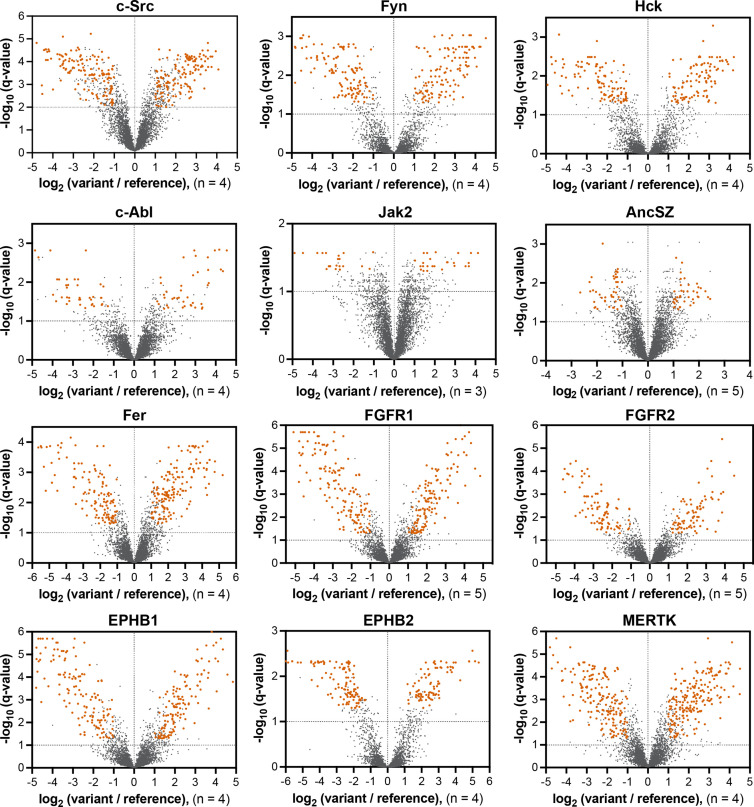
(A) Distribution of enrichment scores from pTyr-Var screens with 13 tyrosine kinases. Each point represents a peptide sequence in the pTyr-Var library. Data points in orange-red represent sequences without a Tyr residue and data points in dark gray represent sequences with a Tyr residue. Each dataset represents the average of three to five replicates. (B) Correlation between enrichment scores and measured phosphorylation rates for 12 peptides (100 μM) with c-Src (500 nM). (C) Correlation between the magnitude of mutational effects for 6 peptide pairs in the pTyr-Var library with mutational effects measured using an in vitro kinetic assay. Error bars in panels B and C represent the standard deviation from 3 to 4 rate measurements and four pTyr-Var screens. (D) Matrix of Pearson’s correlation coefficients for all pairwise comparisons between replicate-averaged pTyr-Var datasets for 13 kinases. (E) Volcano plot depicting mutational effects in the pTyr-Var screen with c-Src kinase domain. Data points represent the average of four replicates. Hits are colored orange-red. (F) Percent phosphorylation of SHP2 wild-type, D61V, and D61N (10 μM) after an hour incubation with c-Src, Fyn, and FGFR1 (1 μM). Error bars represent the standard deviation from 2 to 3 measurements.
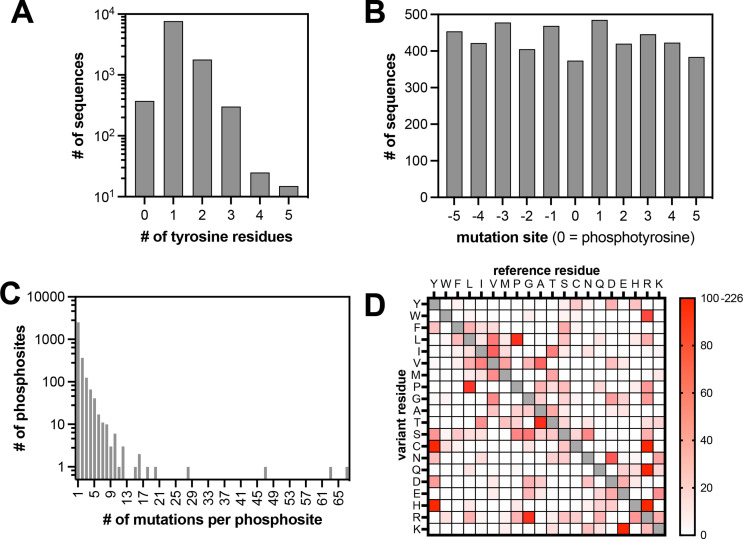
Figure 4—figure supplement 1. Properties of the pTyr-Var library.
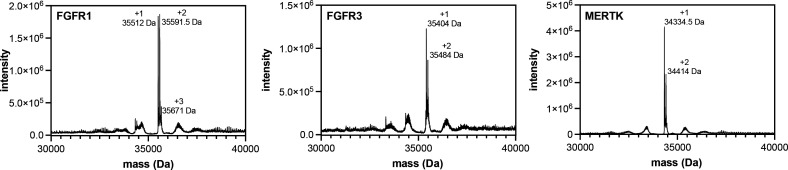
Figure 4—figure supplement 2. Pre-activation of FGFR1, FGFR3, and MERTK by auto-phosphorylation.
Figure 4—figure supplement 3. Matrix of Pearson’s correlation coefficients for all replicates of pTyr-Var screens across all 12 kinases.
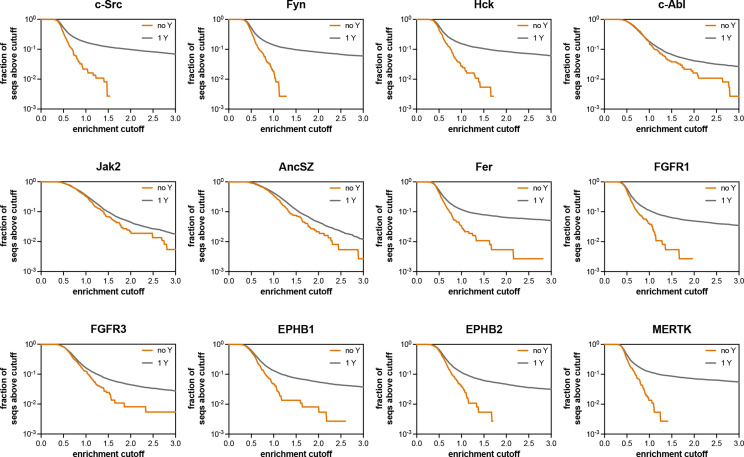
Figure 4—figure supplement 4. Assessment of the extent of enrichment in pTyr-Var screens with 12 kinases.
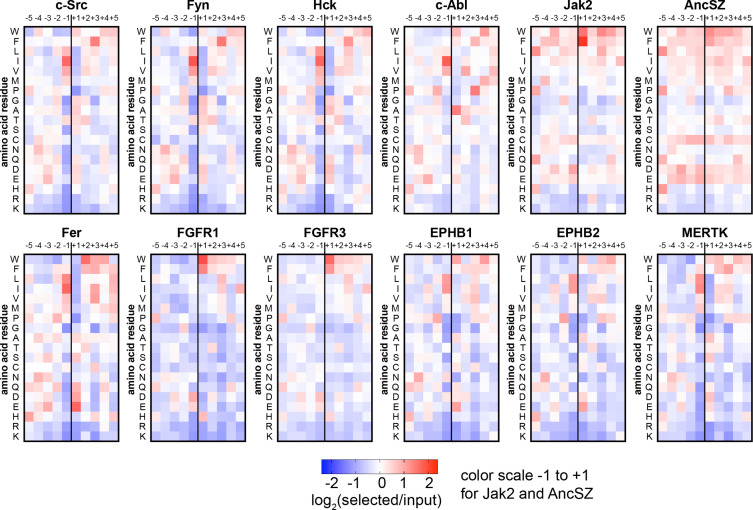
Figure 4—figure supplement 5. Heatmaps depicting the position-specific amino acid preferences for 12 tyrosine kinase domains, extracted from screens with the pTyr-Var library.
Figure 4—figure supplement 6. Volcano plots depicting mutational effects in the pTyr-Var screen for 12 kinase domains.
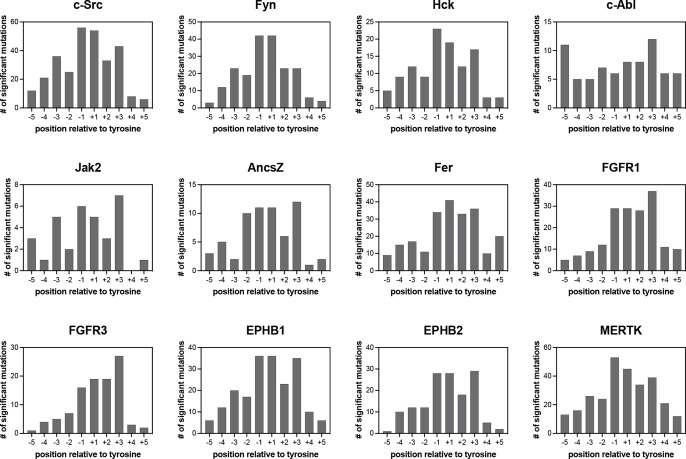
Figure 4—figure supplement 7. Number of significant mutations for each kinase at each position surrounding the central tyrosine residue.
Figure 4—figure supplement 8. Enrichment scores from pTyr-Var screens for phosphorylation of the RET Tyr 981 reference and variant (R982C) peptides by 12 tyrosine kinases.
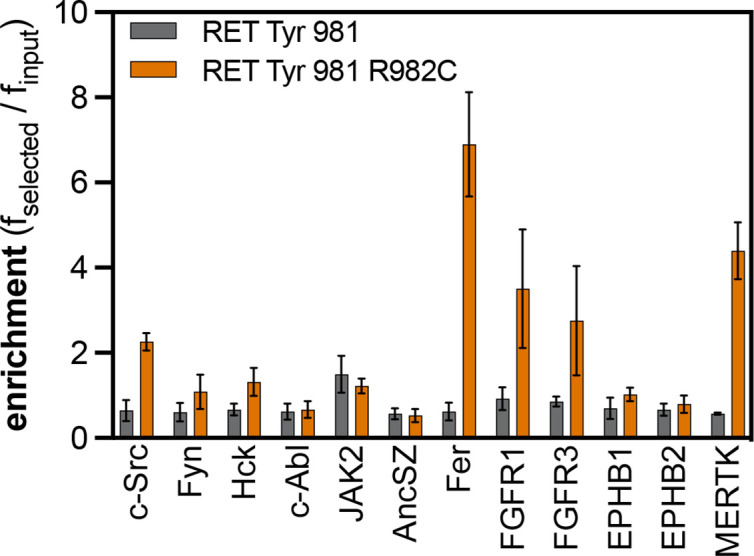
Figure 4—figure supplement 9. Enrichment scores from pTyr-Var screens for phosphorylation of SHP2 Y62 reference and variant (D61N and D61V) peptides by c-Src, Fyn, and FGFR1.