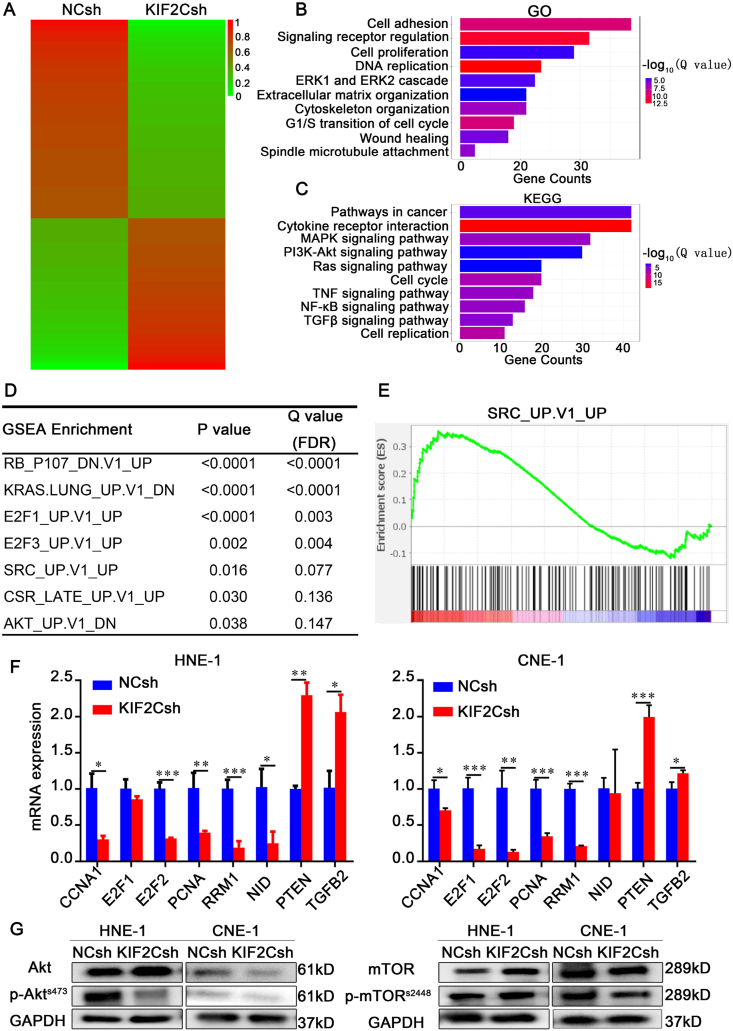
Figure 7.
Downstream genes and pathways regulated by KIF2C. (A) RNA-seq analyses for KIF2C knockdown and its control HNE-1 cells. Heatmap shows the differentially expressed genes with fold change of expression more than 2.0. (B, C) GO enrichment and KEGG pathway analysis based on the differentially expressed genes between the KIF2C knockdown (KIF2Csh) and its control (NCsh) HNE-1 cells. (D) Top gene sets enriched by GSEA analysis. FDR, false discovery rate. (E) GSEA plot of the SRC oncogenic signature (SRC_UP.V1_UP) correlated with KIF2C depletion. (F) Verification of important KIF2C-regulated genes by qRT-PCR. (G) Detection of mTOR and AKT pathways in KIF2C knockdown HNE-1 and CNE-1 cells by immunoblotting.