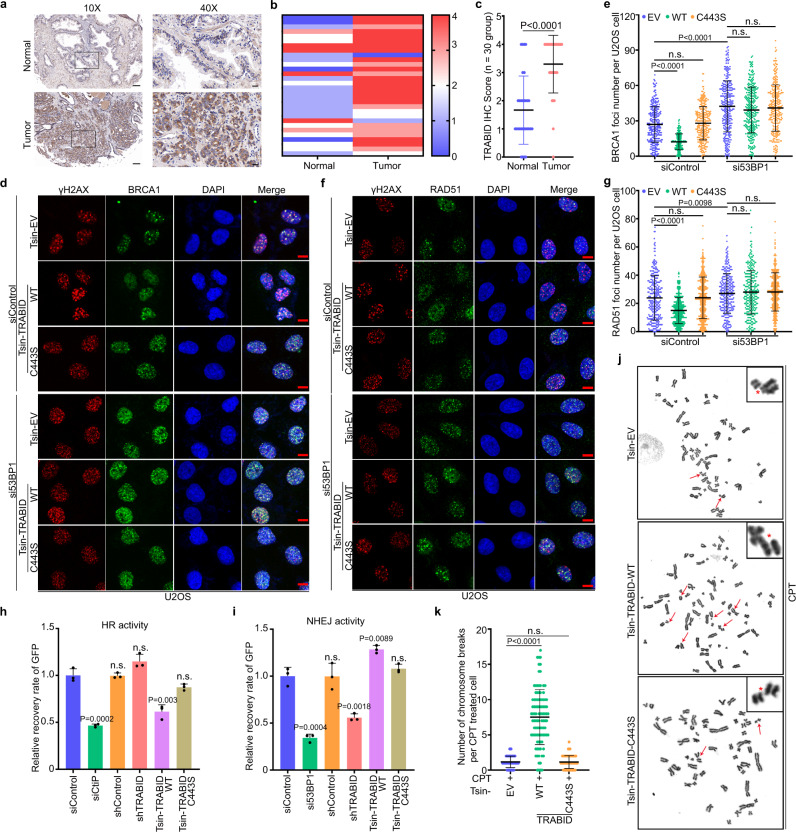
Fig. 5. TRABID overexpression inhibits HR activity and promotes chromosomal instability.
a–c Representative images of IHC staining (a) of TRABID protein on paired prostate cancer patient specimens (n = 30). Scale bar in 10 X fields: 100 μm; Scale bar in 40 X fields: 20 μm. Heat map showing TRABID IHC score in normal and paired tumor tissues (see calculation details in Methods) (b). Quantification of IHC score (c). Data were shown as the mean ± SD of three independent experiments (n = 3). Two-tailed unpaired Student’s t-test. P = 5.3E-7. d–g U2OS cells infected with lentivirus expressing EV, TRABID-WT, or TRABID-C443S and treated with siControl or si53BP1 were exposed to IR followed by IFC of BRCA1 (d) and RAD51 (f) at 1 h after IR. Scale bar, 10 µm. The average foci number (e, g) in each cell were quantified. Data were presented as means ± SD of more than 300 cells from three biological replicates. Two-tailed unpaired Student’s t-test. n.s. not significant. P values based on the order of appearance: e (2.5E-54, 0.3363, 1.9E-22, 0.0659, and 0.4015), g (1.9E-16, 0.9275, 0.0098, 0.4221, and 0.3285). h, i PC-3 cells were transfected with HR or NHEJ reporter in combination with TRABID WT/C443S or small interfering RNA (siRNA) for CtIP or 53BP1 and HR (h) and NHEJ (i) activities were measured. Data were represented as means ± SD of three biological replicates. Two-tailed unpaired Student’s t-test. n.s. not significant. GFP green fluorescent protein. P values based on the order of appearance: h (0.0002, 0.9571, 0.0711, 0.003, and 0.0537), i (0.0004, 0.9819, 0.0018, 0.0089, and 0.2552). j, k PC-3 cells infected with lentivirus expressing EV, TRABID-WT, or TRABID-C443S were treated with CPT (1 μM) for 24 h. Cells were harvested for karyotyping, and chromosome breaks in more than 80 cells from three biological replicates in each group were counted and quantified. Representative images were shown in (j) and quantitative data are shown in (k). Data were shown as the mean ± SD of three independent experiments (n = 3). Two-tailed unpaired Student’s t-test. P values based on the order of appearance:1.3E-37, 0.9348. n.s. not significant. Source data are provided in this paper.