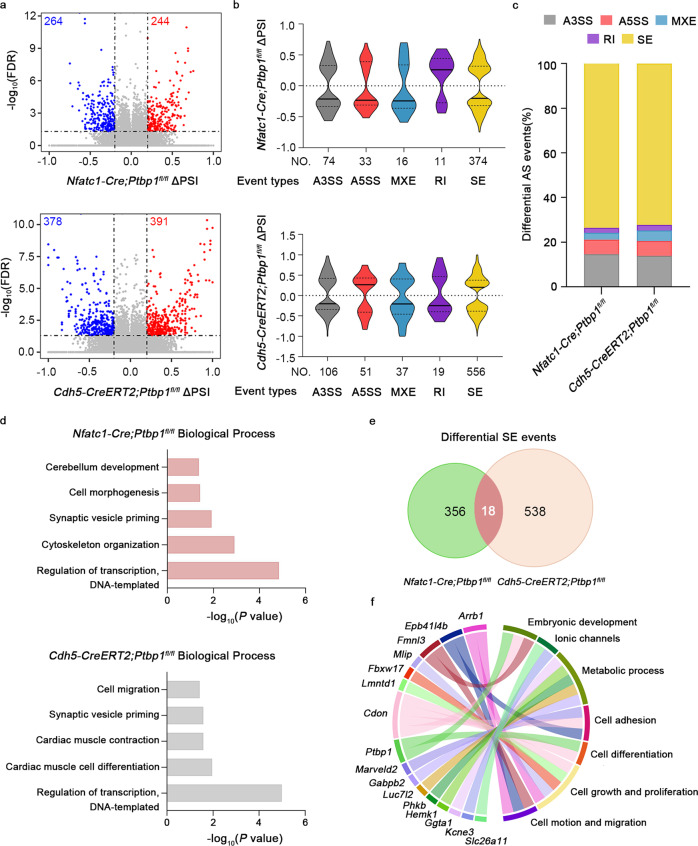
Fig. 6. PTBP1 regulates alternative splicing events related to cardiac development.
a Volcano plots of the differential alternative splicing events identified in E13.5 Nfatc1-Cre;Ptbp1fl/fl hearts and Cdh5-CreERT2;Ptbp1fl/fl hearts compared with Ptbp1fl/fl hearts, respectively. Significant alternative splicing events (FDR < 0.05 and | ΔPSI | ≥ 0.2) are colored as blue and red dots if Ptbp1 deletion results in skipping or inclusion, respectively. n = 2–4 mice per group. PSI percent spliced in. b Violin plot of the distributions of differential alternative splicing event types in Nfatc1-Cre;Ptbp1fl/fl and Cdh5-CreERT2;Ptbp1fl/fl hearts, respectively. A3SS alternative 3’ splice site, A5SS alternative 5’ splice site, MXE mutually exclusive exon, RI retention intron, SE skipped exon. c Stacked bar graph showing the proportions of different types of differential alternative splicing events in Nfatc1-Cre;Ptbp1fl/fl and Cdh5-CreERT2;Ptbp1fl/fl hearts, respectively. d Gene Ontology (GO) enrichment analysis of genes with differential skipping exon (SE) events in Nfatc1-Cre;Ptbp1fl/fl and Cdh5-CreERT2;Ptbp1fl/fl hearts, respectively. GO analysis were performed using R package clusterProfiler. e Venn diagram showing the common significant SE events between Nfatc1-Cre;Ptbp1fl/fl and Cdh5-CreERT2;Ptbp1fl/fl hearts. Exons are required to change in the same direction to appear at the intersection of the two sets. f Circular plot showing the selected functional categories of common genes with differential SE events in E13.5 Nfatc1-Cre;Ptbp1fl/fl and Cdh5-CreERT2;Ptbp1fl/fl hearts. Source data are provided as a Source Data file.