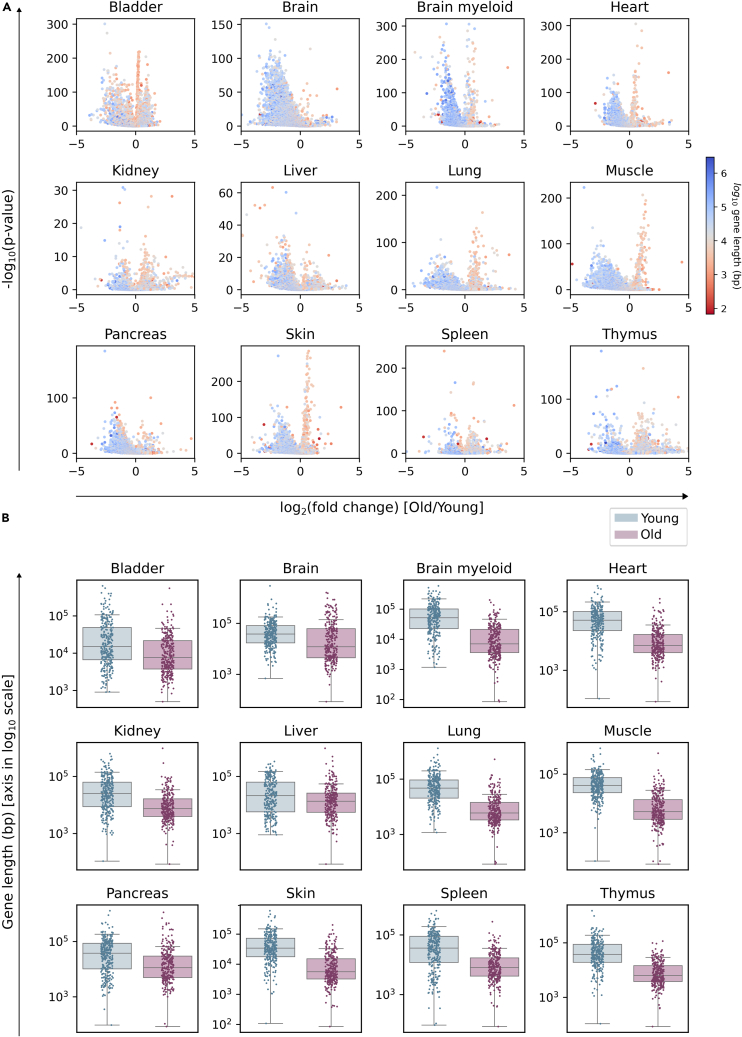
Figure 2.
Differentially expressed genes between young and old individuals show a preferential bias for the downregulation of long genes
Output of the differential expression analysis between old (24 months) and young (3 months) male mice in 12 datasets from the Tabula Muris Senis (using the Wilcoxon method).
(A) Volcano plots showing the length of DEGs: -log10(p value) against log2(fold change). Each gene is colored according to its log10-transformed length.
(B) Boxplots showing the top 300 DEGs for each category. Whiskers extend to extend to the furthest datapoint within the 1.5∗IR (interquartile range). “Young”: top 300 most overexpressed genes in young cells with respect to old cells (blue). “Old”: top 300 most overexpressed genes in old cells with respect to young cells (pink). The difference in DEG length is significant in all tissues (p value < 0.001); see Table S3.
Se also Figures S3 and S4.