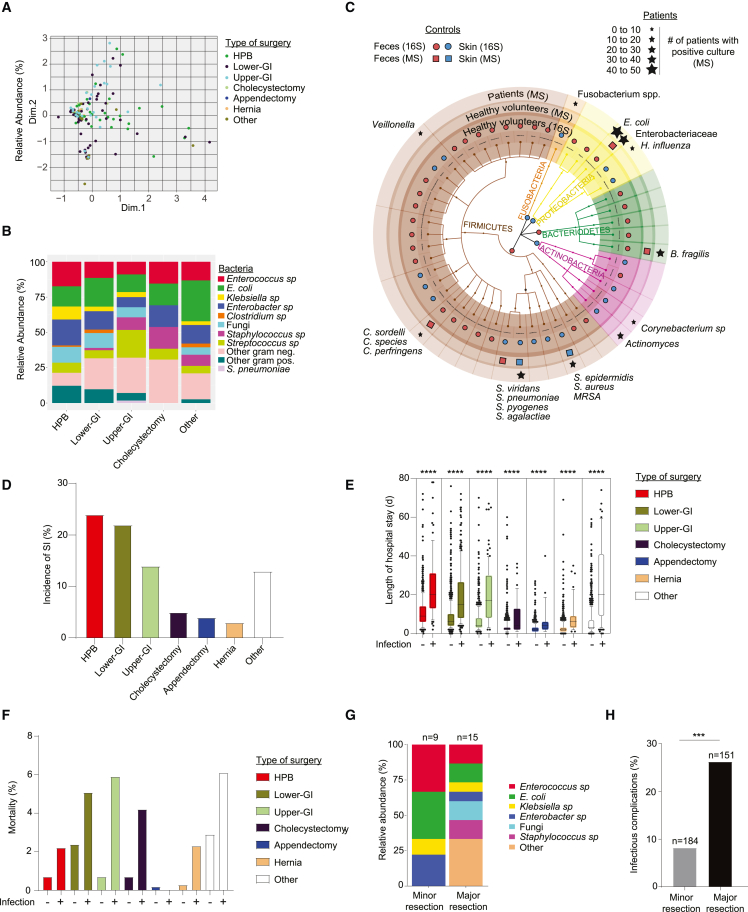
Figure 1.
A multicenter cohort revealed a limited but not surgery-specific composition of intestinal microbes causing surgical infections
A multicenter cohort of 3,515 patients from three Swiss surgical departments was analyzed.
(A and B) Microbial profile of surgical infections (SIs) found in patients with infectious complications. (A) Multiple correspondence analysis (MCA) plot of bacteria found in different surgical procedures. One dot represents one patient. (B) Distribution of bacterial species in SI cultures that underwent different surgical procedures as indicated.
(C) Overall representation of taxa significantly associated with either 96 fecal or 88 skin samples from the healthy volunteers analyzed by 16S rRNA amplicon sequencing are plotted and overlapped with bacterial strains that were identified with MALDI-TOF after culturing (mass spectrometry [MS]) of the fecal or skin samples from 10 healthy volunteers. The red closed circles (16S) or squares (MS) show taxa with increased abundances in fecal samples, and the blue circles/squares show taxa with increased abundances in the skin samples. Stars on top of the graph represent the identified culturable bacteria in SI in patients.
(D) Incidence of SI in different surgical procedures.
(E) Length of hospital stay and type of surgery for patients with or without SI. Each dot represents one patient. Errors bars indicate the median ± SD.
(F) Percentage of mortality related to each type of surgery based on the incidence of infection.
(G and H) A second cohort of 335 patients from the University Hospital of Vienna (Austria) undergoing minor or major liver surgery for colorectal liver metastasis was analyzed. (G) Identification of bacteria cultured from blood of patients with sepsis. (H) Frequency of SI in patients that underwent minor or major liver surgery. HPB, hepatopancreatobiliary surgery; GI, gastrointestinal. Normalized values were analyzed by Student’s t test to compare two experimental groups or by ANOVA to compare more than two groups in parallel. The p values are indicated as follows: ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.