Figure 1.
IL-10 impacts chromatin and transcription in DCs
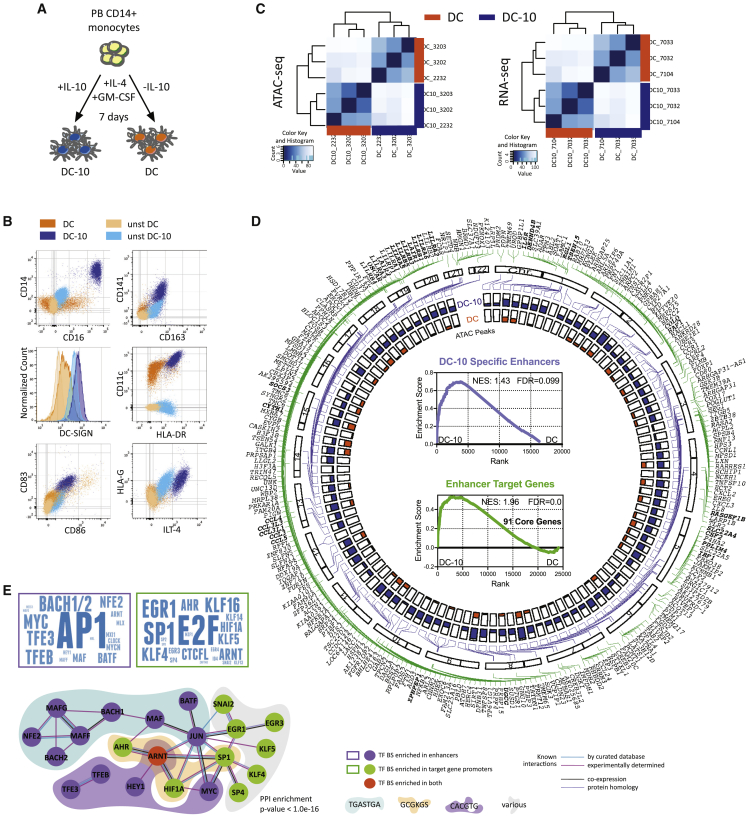
(A) Experimental design. Peripheral blood (PB) CD14+ monocytes were differentiated for 7 days in presence of GM-CSF and IL-4, with (DC-10) or without (DC) IL-10.
(B) Representative plots of DC and DC-10 phenotype by flow cytometry (FC).
(C) Unsupervised clustering heatmaps of ATAC-seq (left) and RNA-seq (right) data from DC and DC-10 from three independent donors.
(D) Circoplot depicting the following: outer layer, location of FANTOM5-identified DC-10-specific enhancers (purple lines), and enhancer target genes (green lines) along chromosomes (bold genes = targets of multiple enhancers); inner layer, ATAC broadpeak maximum value for each enhancer in DC-10s (blue) and DCs (orange). Inner core: GSEA analysis of DC-10-specific enhancers (top, purple) and enhancer target genes (bottom, green) in DC-10 vs. DC transcriptome. Normalized enriched score (NES), false discovery rate (FDR), and number of core genes (bottom) are indicated.
(E) Transcription factor (TF) binding predictions. Left, word clouds displaying TFs whose binding sites (BSs) were significantly enriched in the 107 DC-10 enhancers (top, purple) and 268 DC-10 enhancer target protein-coding gene promoters (bottom, green); size of TF is proportional to its enrichment score from motif finder analysis. Right, protein-protein interaction network built on TF from word clouds; only connected nodes are shown. Colored clouds identify TFs with shared BSs, with consensus site indicated below. PPI, protein-protein interaction. See Figure S1.