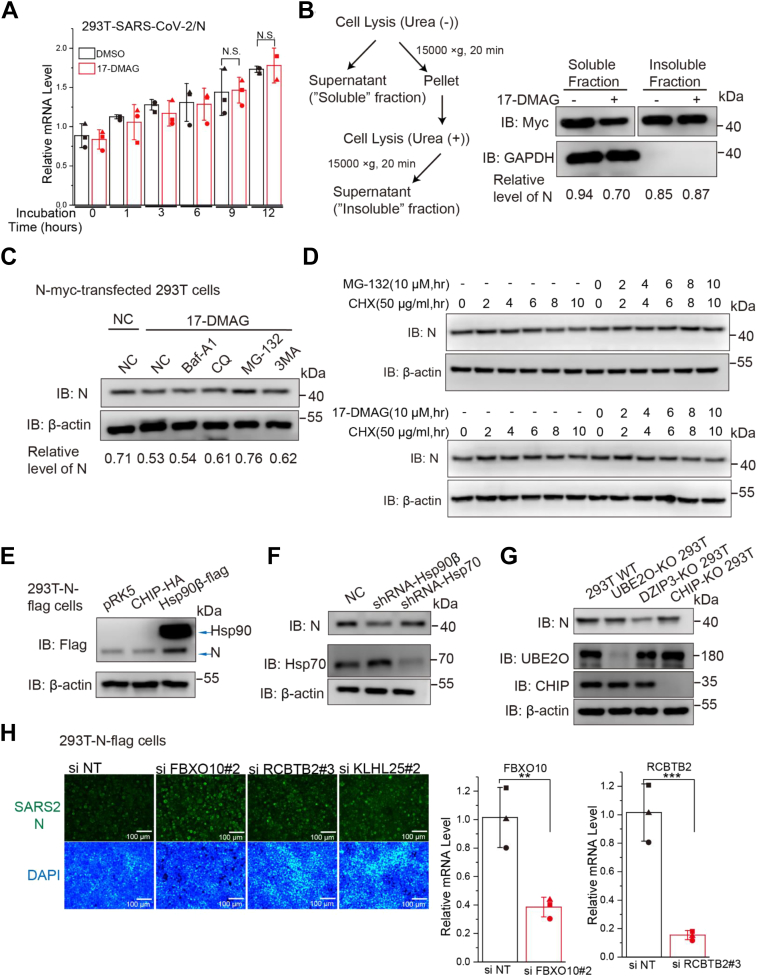
Figure 4.
Hsp90 inhibition mediates proteasomal degradation of viral N protein.A, effect of 17-DMAG treatment on viral N mRNA levels, as determined by qRT-PCR; ∗p ≤ 0.05; ∗∗p ≤ 0.01; n.s.: not significant. B, at 24 h post-transfection, soluble and insoluble fractions of the cells were prepared as indicated and subjected to immunoblot, the relative level of N protein was quantified by immunoblot scanning and normalized with respect to the amount of GAPDH. C and D, HEK293T cells were transfected with N-myc after blocking protein synthesis with cycloheximide (CHX) and the cells were then treated with the protease inhibitor MG132 or the autophagy inhibitor 3-methyladenine (3MA), chloroquine (CQ), or bafilomycin A1 (Baf A1). The relative level of N protein was quantified by immunoblot scanning and normalized with respect to the amount of β-actin. E, HEK293T-N-flag cells were transfected with CHIP-HA vector, with Hsp90β-flag vector used as positive control. F, HEK293T cells were treated with shRNA-Hsp90β or shRNA-Hsp70 and transfected with N-myc. G, WT or CHIP-KO HEK293T cells were transfected with myc-tagged SARS-CoV-2 proteins. H, HEK293T-N-flag cells were seeded in 24-well plates and incubated with siRNA for 84 h, cells were immunostained with anti-N (green) and nuclei were stained with DAPI (blue). The cells were observed by fluorescence microscopy. Intracellular FBXO10 and RCBTB2 mRNA levels were measured by real-time RT-PCR. The data were first normalized to cellular GAPDH mRNA and then to negative control siRNA–treated samples to obtain fold induction. Data are shown as the mean ± SD from three independent experiments. CHIP, C terminus of Hsp70-interacting protein; Hsp90, heat shock protein 90; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.