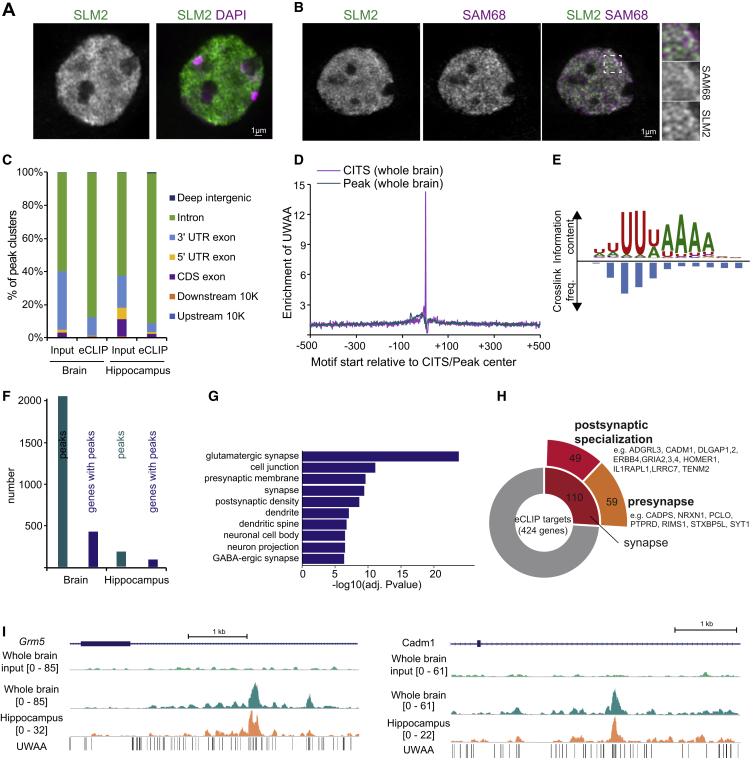
Figure 1.
SLM2-bound mRNAs encode synaptic proteins
(A) Endogenous SLM2 immunoreactivity (green) and DAPI (purple) in hippocampus. Scale bar, 1 μm.
(B) Immunostaining for SLM2 (green) and SAM68 (magenta) in cultured hippocampal neurons (day in vitro 12). Insets show enlargement of boxed area. Scale bar, 1 μm.
(C) Cluster annotation summary for input and eCLIP samples.
(D) Enrichment of UWAA around cross-link-induced truncation site (CITS), calculated from frequency of UWAA starting at each position relative to the inferred cross-link sites, normalized by frequency of the element in flanking sequences in whole-brain eCLIP data. Enrichment of UWAA around the CLIP tag cluster peak center shown for comparison.
(E) SLM2 binding motif determined by mCross in whole-brain eCLIP data. Cross-linking frequencies at each motif position represented by blue bars.
(F) Number of high-confidence SLM2 targets identified by CLIPper and IDR analysis (log2 fold change ≥ 2 and −log10(IDR) ≥ 2) in whole-brain and hippocampal eCLIP samples.
(G) Gene Ontology analysis (DAVID tools) of genes with significant SLM2 binding sites (whole brain). Top 10 enriched Gene Ontology categories for cellular compartment displayed.
(H) Sunburst chart and gene examples associated with synaptic function of high-confidence eCLIP targets from whole-brain samples identified by CLIPper and IDR.
(I) SLM2 eCLIP read densities on eCLIP targets compared with size-matched input. UWAA motif enrichment in whole-brain (green) and hippocampal (orange) samples. Coordinates shown are Grm5 chr7: 87,601,936–87,607,378 and Cadm1 chr9: 47,836,291–47,840,954.