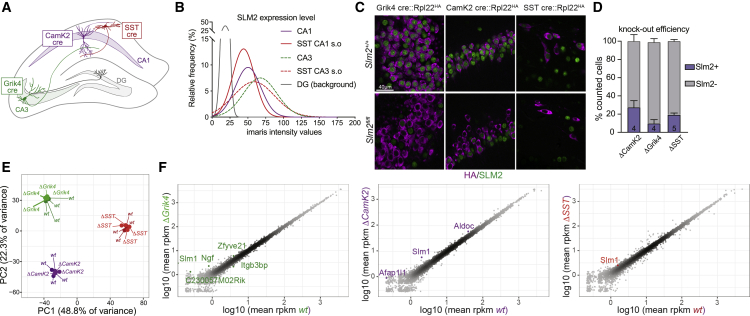
Figure 2.
Conditional ablation of SLM2 in hippocampal cell types
(A) Cre drivers to assess the molecular profile of hippocampal Cornu Ammonis (CA) 1 (CamK2-cre), CA3 (Grik4-cre), and SST-positive (SST-cre) neurons.
(B) Fitted Gaussian curves of relative frequency of SLM2 immunoreactivity in CA1, CA3 and SST cre-positive neurons in the stratum oriens (s.o) of CA1 and CA3. Background immunoreactivity defined based on staining of global Slm2KO mice. n = 3 animals each. CA1: 68 cells, CA3: 75 cells, SST CA1 s.o: 73 cells, SST CA3 s.o: 60 cells, dentate gyrus (DG): 62 cells.
(C) SLM2 expression in cre-positive cells defined by immunoreactivity for conditional Rpl22HA allele in Slm2+/+ and Slm2fl/fl mice (HA: magenta, SLM2: green, scale bar, 40 μm).
(D) Quantification of SLM2 deletion efficiency at P42–45 in CA1 (ΔCamK2, N = 4, n = 1,081) and CA3 (ΔGrik4, N = 4, n = 1,070) and at P16–18 in SST (ΔSST, N = 5, n = 157) neurons. Mean ± SEM.
(E) Principal-component analysis of genes expressed in hippocampal Slm2 wild-type and conditional knockout RiboTRAP pull-downs (wild type [WT] in green: Grik4cre:Rpl22HA/HA N = 4, ΔGrik4: Grik4cre::Rpl22HA/HA::Slm2fl/fl N = 4; WT in purple: Camk2cre::Rpl22HA/HA N = 3, ΔCamK2: Camk2cre::Rpl22HA/HA::Slm2fl/fl N = 3; WT in red: SSTcre::Rpl22HA/HA N = 4, ΔSST: SSTcre::Rpl22HA/HA::Slm2fl/fl N = 4). Variances explained by principal component 1 (PC1) and PC2 are indicated. Variance stabilization transformation was utilized to normalize gene expression.
(F) Correlation analysis of the mean log10 transformed, normalized transcript counts (reads per kilobase million [rpkm]) between WT (x axis) and mutants (y axis). Differentially expressed genes (fold change ≥ 1.5, adjusted p value Benjamini and Hochberg ≤ 0.05) are marked in color.