Figure 3.
SLM2 directly regulates alternative splicing of mRNAs encoding synaptic proteins
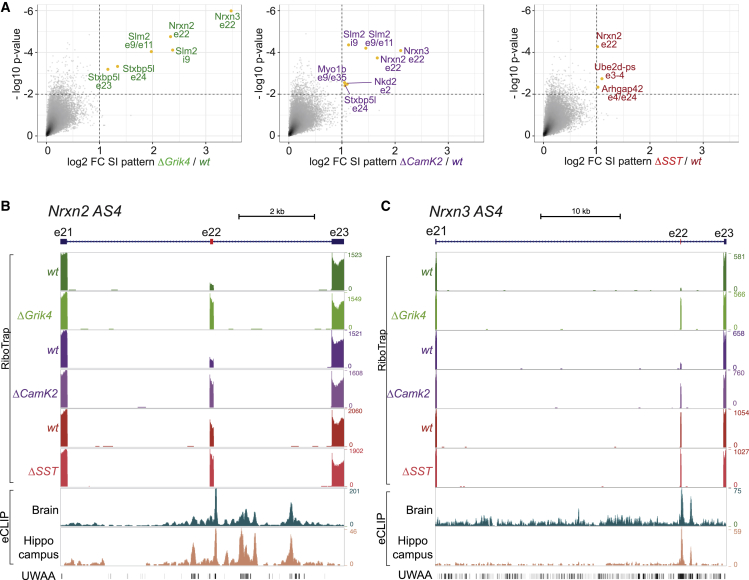
(A) Log2 fold change splicing index (FC SI) and −log10 p values of splicing patterns comparing WT and Slm2 mutants. Significantly regulated exons are marked in color (fold change ≥ 2, p ≤ 0.01).
(B and C) Sequencing tracks for WT and mutant (Δ) RiboTrap samples for hippocampal Grik4, CamK2, and SST cells and eCLIP analysis in whole brain and hippocampus for de-regulated exons of Nrxn2 and Nrxn3. SLM2 binding events in downstream introns align with UWAA motifs. Coordinates: Nrxn2 chr19:6,509,778–6,517,248; Nrxn3 chr12:90,168,920–90,204,818.