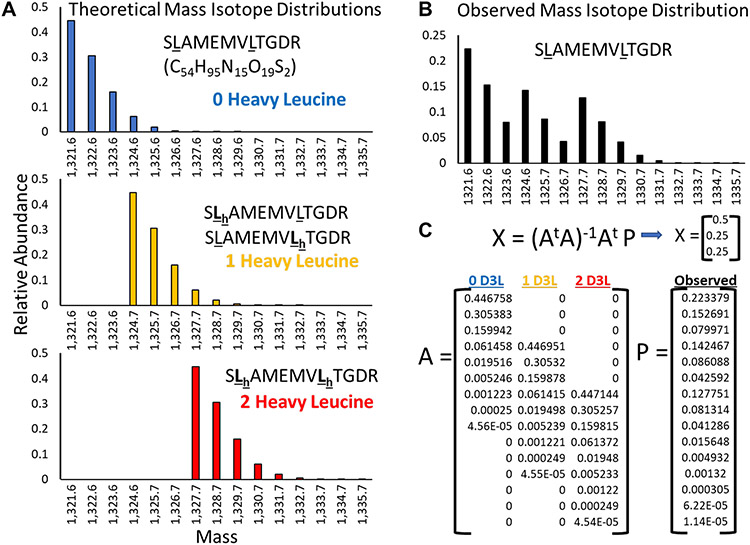
Figure 4.
Correction for naturally occurring heavy isotope signals in TurnoveR. Based on the molecular formula and the natural proportion of heavy isotopes of carbon, hydrogen, nitrogen, oxygen, and sulfur, the theoretical isotope envelopes of every peptide are calculated for every leucine-containing peptide isotopologue (every permutation of light/heavy leucine). (A) For example, the theoretical isotope envelopes of SLAMEMVLTGDR containing zero, one, or two heavy leucines are shown. The isotope envelopes have overlapping mass. (B) In a theoretically observed distribution of SLAMEMVLTGDR, 50% of total MS1 intensity was contributed by the unlabeled peptide isotopologue, 25% from the singly labeled peptide isotopologue, and 25% from the fully labeled peptide isotopologue. (C) Matrix math is used to solve for the relative contribution of signal from each peptide isotopologue. Matrix A is defined by the fractional intensity of each isotopologue (columns) at different molecular masses (rows). Matrix P is the observed intensity at each molecular mass.