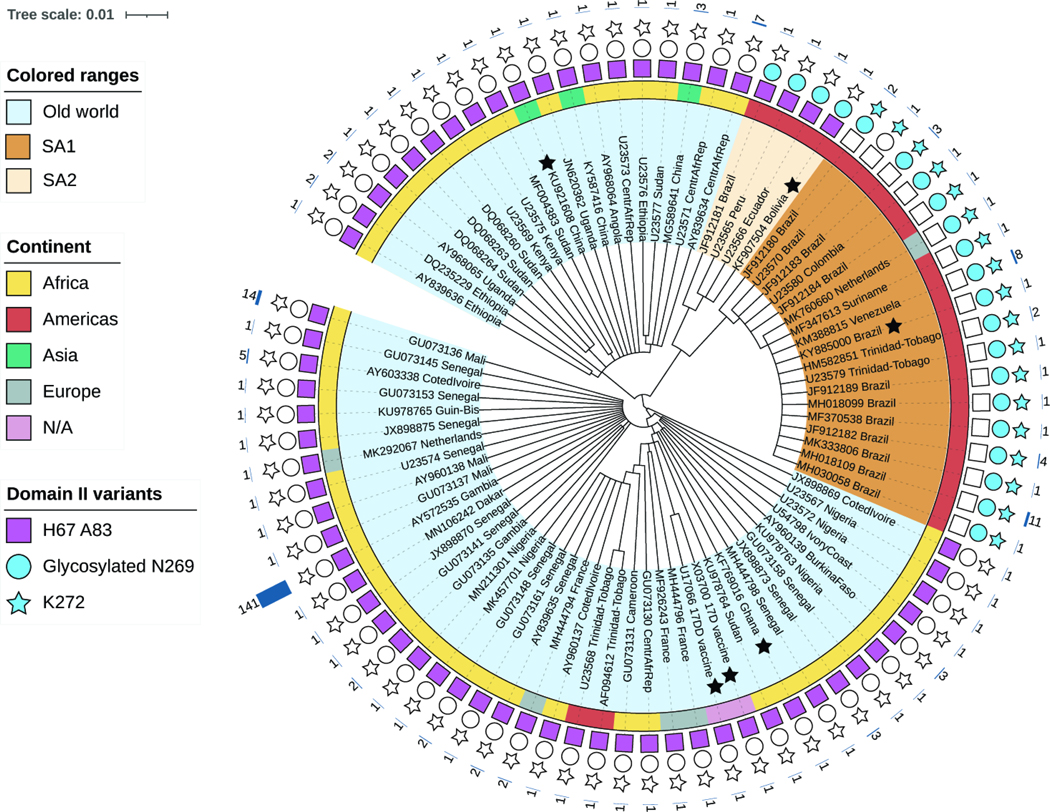
Figure 7. Genetic makeup of YFV strains at polymorphic sites 1 and 2.
Amino acid-based tree depicting E proteins from representative YFV strains (scale bar represents the number of amino acid substitutions per site). Colored ranges denote viral origin and clade(s): i) Old World (light blue), ii) SA1 (South American genotype 1, brown). SA2 (South American genotype 2, beige). Black stars indicate YFV sequences used in this study. The inner ring is color-coded according to the continent of origin. Note that all sequences of apparent Asian or European origin are ex-African (if clustering with Old World) or ex-South American (if clustering with SA1). N/A, not applicable. Sequence features are represented with a square (Site 1: H67 A83), circle (Site 2a: predicted glycan at N269), or star (Site 2b: K272). Features present in sequence are represented with filled shapes. The outer char bar denotes the number of strains being represented at each branch of the tree.