FIGURE 1.
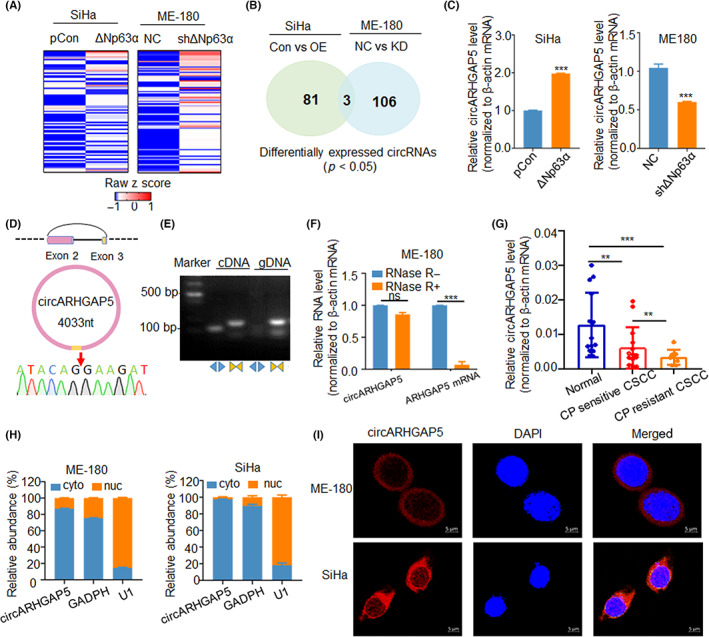
Identification and characterization of circular RNA ARHGAP5 (circARHGAP5) in cervical squamous cell carcinoma (CSCC) cells and tissues. (A) Heatmap of circRNA expression profile in SiHa/Con with SiHa/ΔNp63α and ME‐180/NC with ME‐180/shΔNp63α stable cell lines. (B) Overlap of 193 differentially expressed circRNAs in these two groups of cells. (C) Verification of circARHGAP5 expression levels in the above two groups of cells. (D) Sanger sequencing of backsplice junction site for circARHGAP5. (E) PCR analysis for circARHGAP5 in cDNA and genomic DNA (gDNA) with divergent and convergent primers. (F) Relative RNA levels of circARHGAP5 and ARHGAP5 with or without RNase R digestion. (G) Expression level of circARHGAP5 in normal cervical tissues and untreated and cisplatin (CP)‐resistant CSCC tissues. (H) Nuclear (nuc) and cytoplasmic (cyto) levels of circARHGAP5 in ME‐180 and SiHa cells. GADPH served as cytoplasmic control, while U1 served as nuclear control. (I) FISH assay of circARHGAP5 (red) in ME‐180 and SiHa cells. The location of circARHGAP5 (red) in SiHa and ME‐180 cells was determined by FISH assay. DAPI‐stained nuclei are blue. Scale bar = 5 μm. **p < 0.01, ***p < 0.001. Con, control; NC, negative control; ns, not significant; OE, overexpression.
