Figure 1. Modelling predicts that force can improve or impair antigen discrimination.
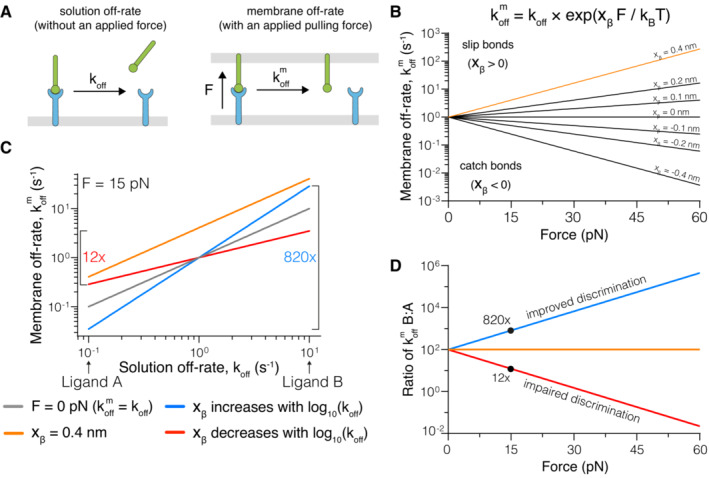
- Depiction of (left) the dissociation of a soluble ligand and its solution off‐rate (koff) and (right) a membrane‐anchored ligand and its membrane off‐rate (km off), where the interaction is exposed to a pulling force (F).
- The dependence of km off on the pulling force at the indicated force sensitivity parameters (xβ) with koff = 1 s−1.
- Dependence of km off on koff at zero force (grey) or under an applied force of 15 pN (coloured lines), when xβ is constant (orange), positively (blue), or negatively (red) correlated with the solution koff. In this example a 100‐fold change in koff (Ligand A vs. Ligand B) can be increased to a ~ 820‐fold or decreased to a ~ 12‐fold change in km off, depending on whether xβ is positively or negatively correlated with the koff, respectively.
- The fold‐change in km off for Ligand B versus Ligand A over the applied force.
Data information: All calculations are performed using the formula in (B) with xβ = 0.4 nm (orange), xβ = +0.3 log10(koff) (blue), and xβ = −0.3 log10(koff) (red) in panels C and D.
