Figure 6. MitoStores form proximity to mitochondria.
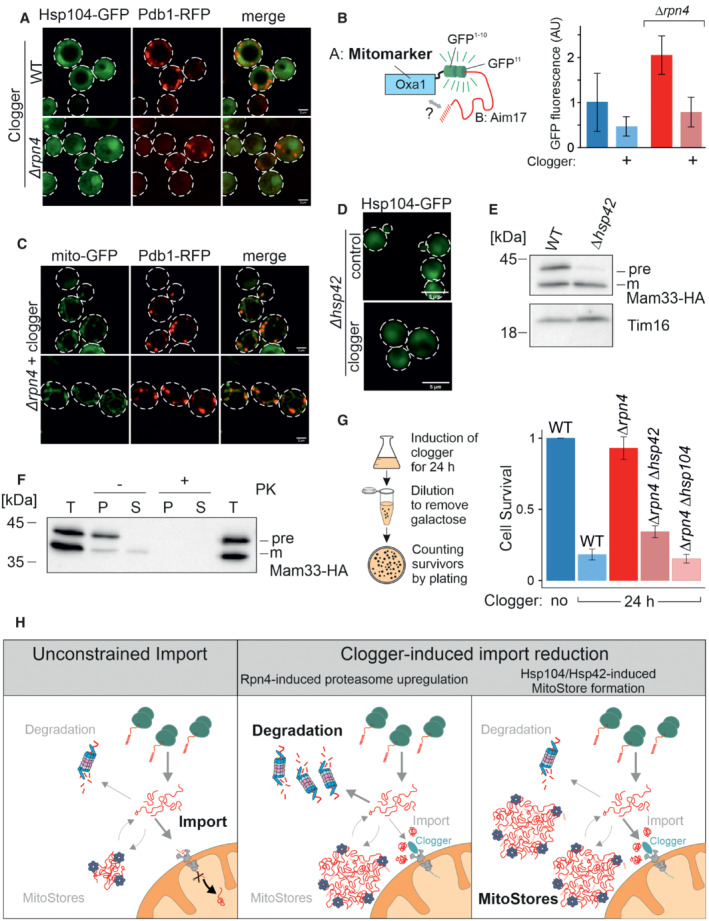
- After 4.5 h of the galactose‐induced expression of clogger and Pdb1‐RFP, cells were incubated for 4 h in the absence of galactose. Whereas Hsp104‐GFP was distributed throughout the cytosol after this chase period, Pdb1‐RFP showed a distribution pattern characteristic of mitochondrial proteins. Scale bars, 2 μm.
- To verify that stored proteins are chased into the mitochondrial matrix, Aim17‐GFP11 was co‐expressed with a clogger for 4.5 h in cells containing mitochondria with Oxa1‐GFP1‐10. Cells were washed in galactose‐free buffer and further incubated for 4 h before fluorescence was measured to quantify the matrix‐localized Aim17 proteins. Data are displayed as mean ± standard deviations from n = 3 independent biological replicates.
- Clogger was expressed for 4.5 h before colocalization of MitoStores (visualized by Pdb1‐RFP), and mitochondria (stained with matrix‐residing mito‐GFP) were assessed. Scale bars, 2 μm.
- Clogger was expressed for 4.5 h in Δhsp42 cells before Hsp104‐GFP was visualized. Please note that in the absence of Hsp42, no aggregates were detected. Scale bars, 5 μm.
- Clogger and Mam33‐HA were co‐expressed for 4.5 h in wild‐type and Δhsp42 cells. Mitochondria were isolated and subjected to Western blotting to detect mature (m) and precursor (pre) forms of Mam33‐HA. The matrix protein Tim16 served as a loading control.
- The mitochondria used for (E) were treated with proteinase K (PK) to remove surface‐exposed proteins. Mitochondrial membranes were lysed with NP‐40 and soluble (S) and aggregated (P) proteins were separated by centrifugation. T, total.
- To measure the toxicity of clogger expression, clogger proteins were induced for 24 h with lactate medium containing 0.5% galactose in the mutants indicated. Aliquots were removed, and the number of living cells was assessed by a plating assay on glucose‐containing plates. The resistance of Δrpn4 to clogger expression depends on Hsp104 and Hsp42. Data are displayed as mean ± standard deviations from n = 3 independent biological replicates.
- Schematic representation of cytosolic MitoStores.
Source data are available online for this figure.
