Figure 4. Structures of the yeast and human SSU processome.
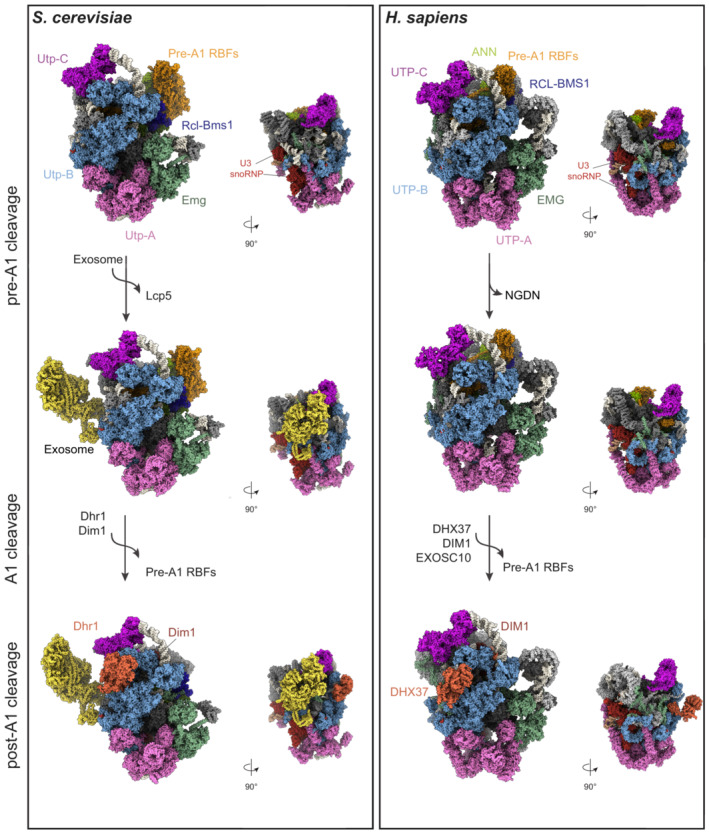
Cryo‐EM structures of the yeast (PDB ID: 6ZQB, 7AJT, and 7AJU) and human (PDB ID: 7MQ8, 7MQ9, 7MQA) SSU processome particles in the pre‐A1, pre‐A1* and post‐A1 cleavage states, showing a comparison of their overall architecture. The conserved RNA components and subcomplexes are color‐coded. The pre‐rRNA (white) and individual sub‐complexes such as UTP‐A (pink), UTP‐B (light blue), UTP‐C (violet), Emg1 (green), ANN (light green) complex, and additional RBFs, are shown as surfaces. KRR1/Krr1, C1orf131/Faf1, and the NAT10/Kre33 module are indicated as pre‐A1‐specific RBFs (sand). They are released after A1 cleavage. The exosome complex (yellow) is bound to the pre‐A1* yeast particle and it is associated only with the human post‐A1 structure. In the post‐A1 structures, DHX37/Dhr1 and DIM1/Dim1 are displayed in orange.
