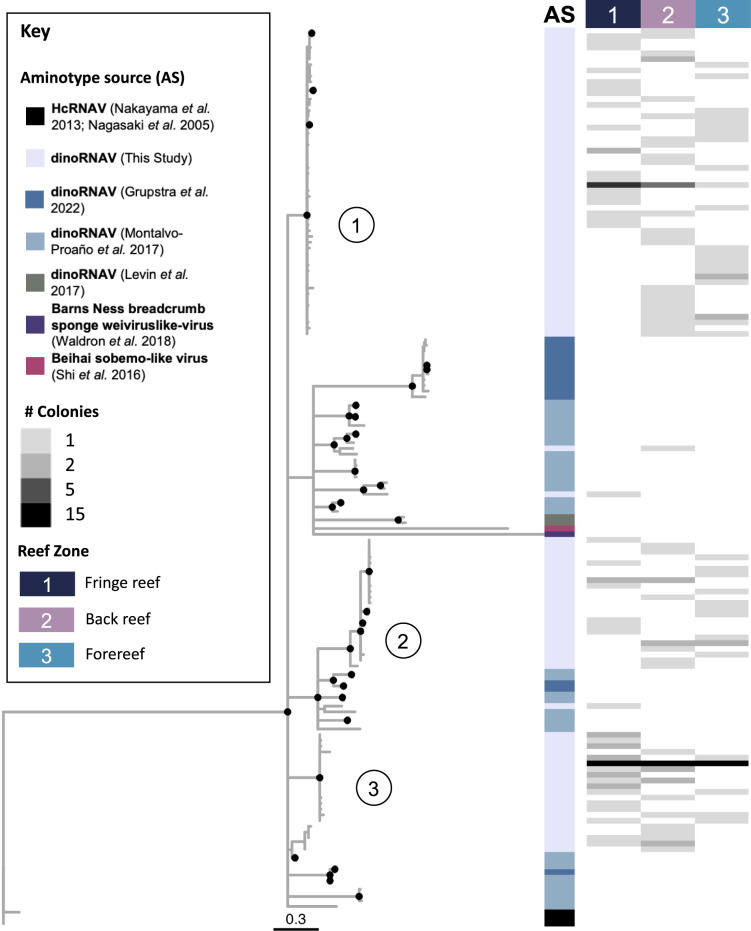
Fig. 3. Maximum likelihood tree of dinoRNAV major capsid protein (mcp) unique amino acid sequences (‘aminotypes’) from Porites lobata-Cladocopium C15 holobionts in this study as well as previously reported dinoRNAV mcp sequences and related viral sequences.
A maximum likelihood phylogeny with 500 bootstrap iterations was generated using dinoRNAV mcp aminotype sequences generated from Porites lobata-Cladocopium C15 holobionts from coral reefs of Moorea, French Polynesia (South Pacific) in this study, as well as previously reported mcp sequences from the Great Barrier Reef [30], Moorea [29], and related viral sequences such as the Beihai sobemo-like virus and the Barns Ness breadcrumb sponge weiviruslike-virus [44, 54]. This tree was outgroup rooted by the longest branch, Heterocapsa circularisquama RNA virus (HcRNAV, [52, 53]). Colors in the vertical bar (Aminotype Source, AS) indicate the origin of a given sequence. Branches with less than 50% bootstrap support were collapsed; branches with bootstrap support >70% are depicted with a black dot. Only aminotypes that were present in >10% relative abundance in at least one sample are included in the phylogeny. Gray tone heat maps indicate the number of colonies from which each aminotype was detected, separated by reef zone (rightmost columns 1–3). Circled numbers reference the three largest clades of dinoRNAV aminotypes from this study.